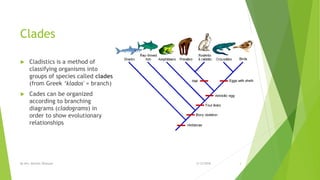
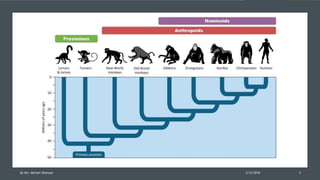
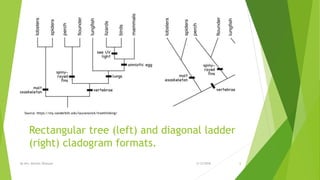
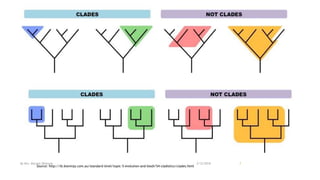
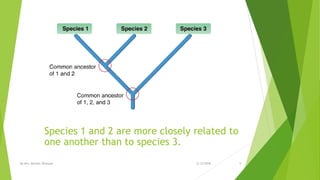
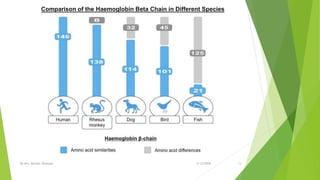
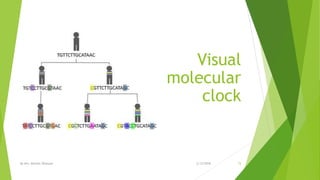
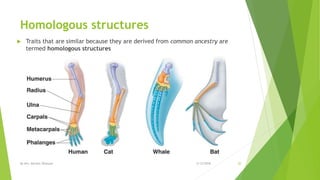
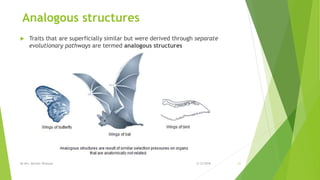
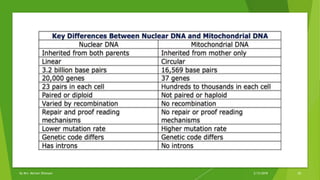
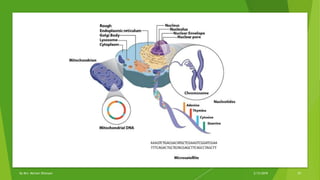
The document discusses cladistics, a method for classifying organisms into clades based on their evolutionary relationships, which is visually represented by cladograms. It highlights the importance of molecular evidence, such as DNA and protein sequences, in determining relatedness and divergence among species. Additionally, it explains concepts such as homologous and analogous structures, convergent evolution, adaptive radiation, and the significance of mitochondrial DNA in tracing evolutionary paths.