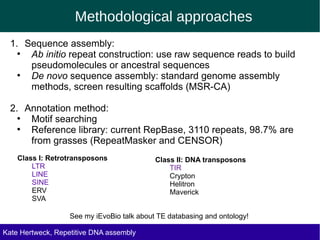
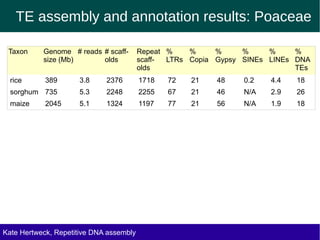
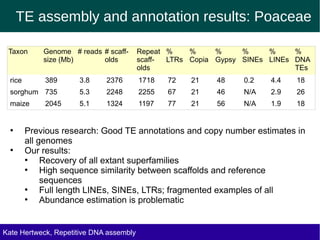
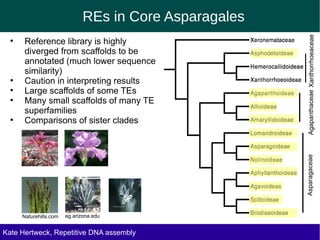
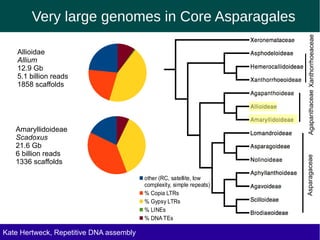
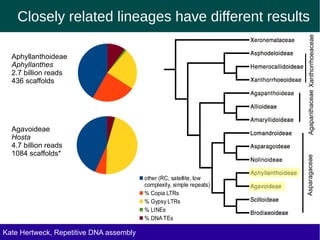
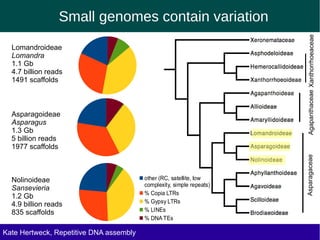
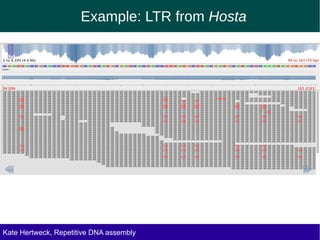
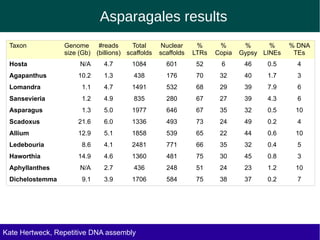
The document discusses methods for assembling and annotating repetitive DNA elements from genome survey sequencing (GSS) data in grasses and other plant groups. In grasses like rice, sorghum, and maize, the authors were able to recover full-length transposable elements and estimate abundance, due to existing reference libraries. However, in the order Asparagales, which contains very large genomes, reference libraries are highly diverged, annotation is more difficult, and abundances may not be accurately estimated. The authors find variation in repetitive DNA content even among closely related lineages in Asparagales.