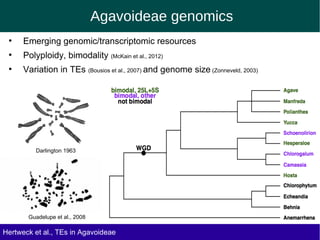
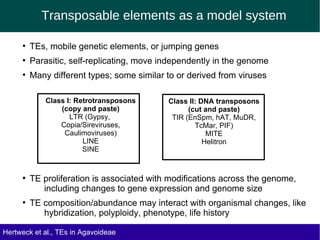
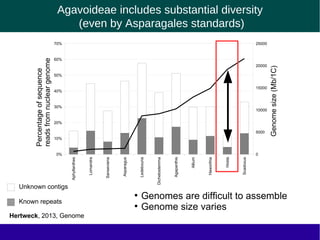
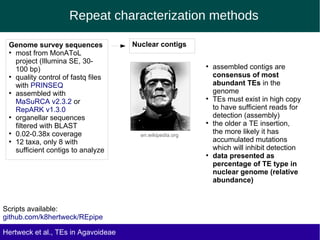
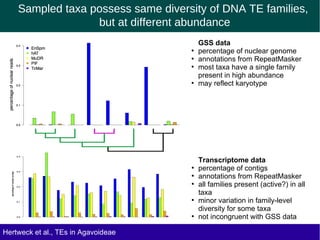
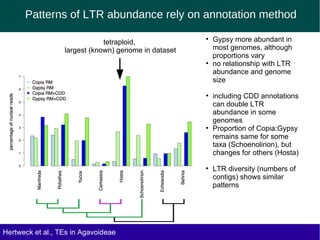
The document discusses the transposable elements (TEs) in the Agavoideae subfamily of Asparagaceae, highlighting their evolutionary significance and economic importance. It outlines the efforts to characterize TEs, their abundance, and diversity across different species, while noting challenges in genome assembly and the influence of TEs on genomic evolution and life history traits. Future research aims to refine TE annotations and explore correlations between transposon composition and genomic evolution patterns.