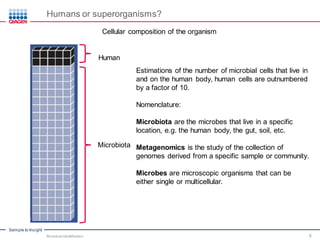
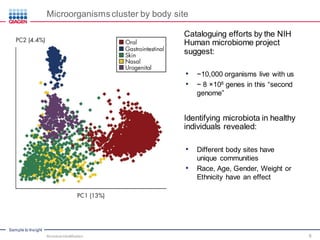
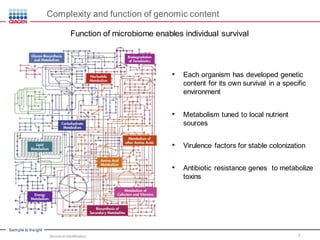
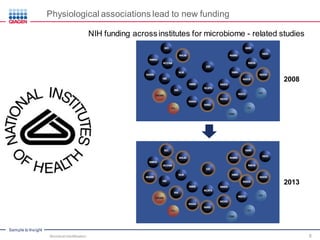
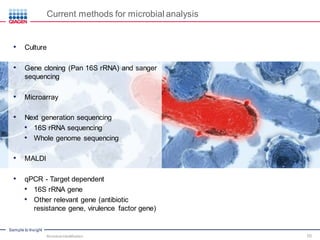
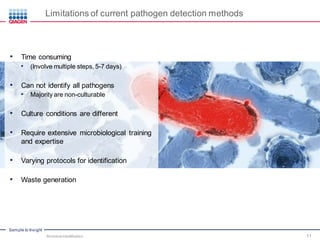
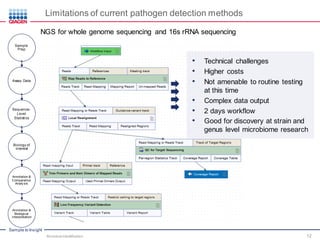
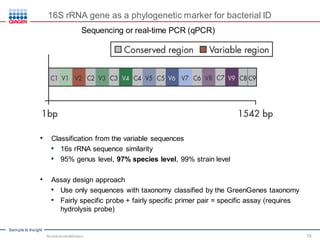
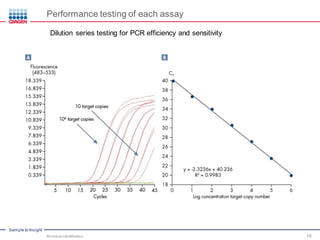
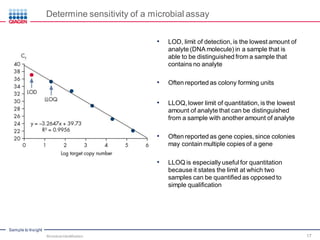
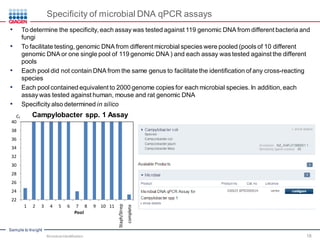
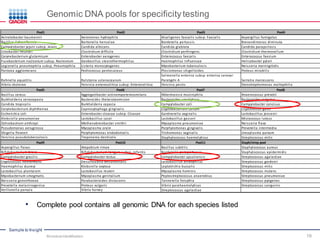
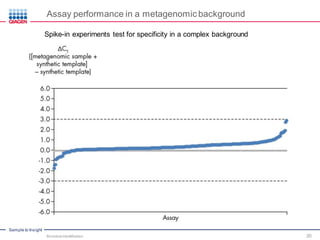
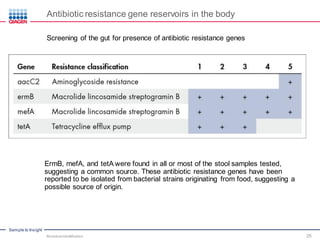
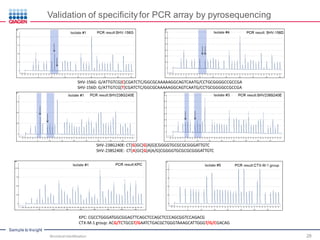
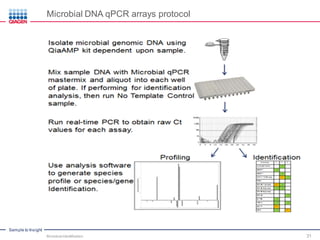
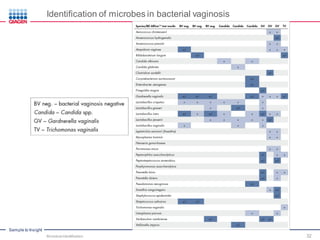
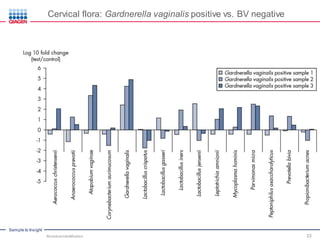
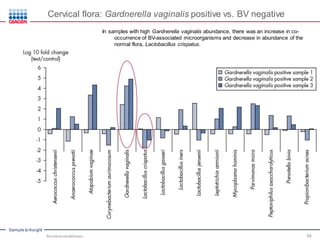
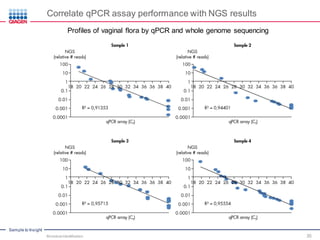
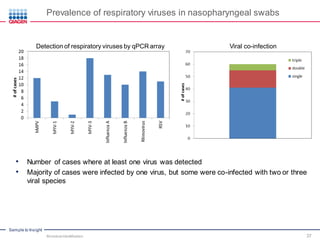
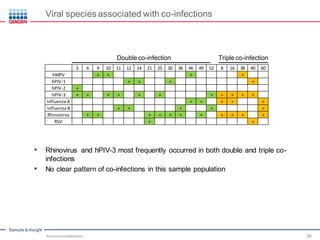
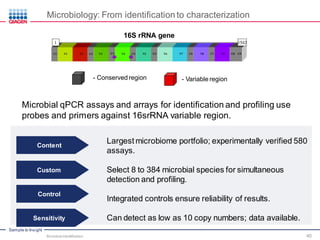
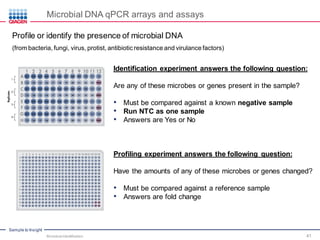
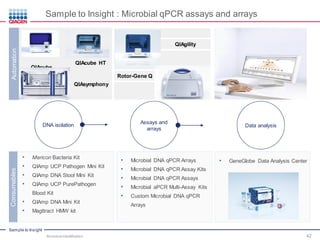
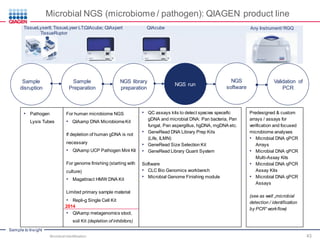
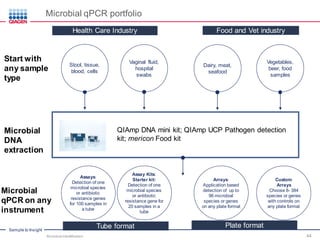
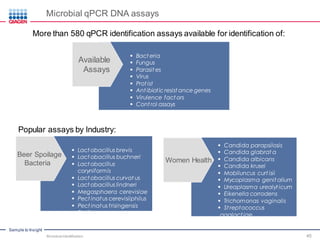
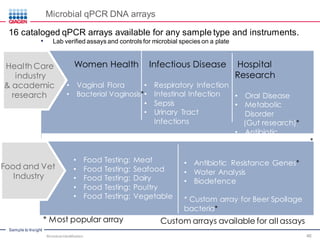
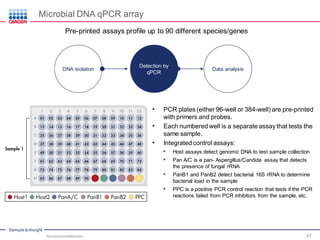
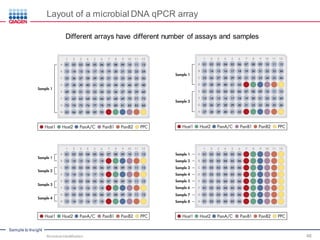
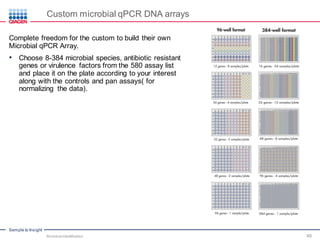
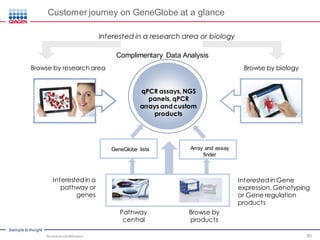
This document discusses the development of rapid detection methods for microbial and microbiome analysis and their applications to human health. It provides an overview of QIAGEN's microbial qPCR products and discusses focused metagenomics applications like screening for antibiotic resistance genes in the food supply and human gut. Limitations of current methods are outlined and the benefits of qPCR for rapid, specific, and sensitive microbial detection are described.