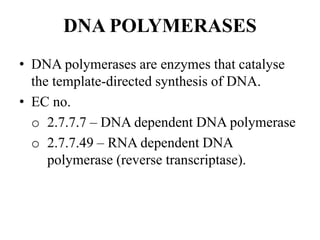
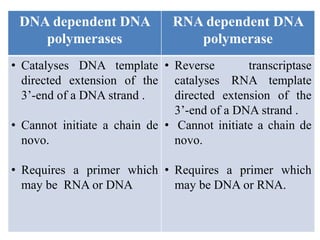
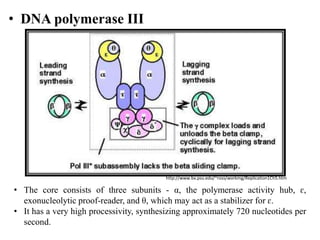
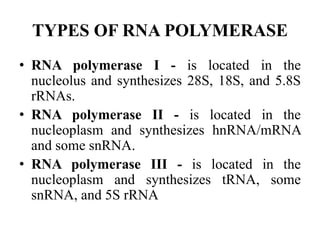
DNA and RNA polymerases are enzymes that catalyze the template-directed synthesis of DNA or RNA. There are several types of both DNA and RNA polymerases that serve different functions. DNA polymerases were first discovered in E. coli in 1955 and help replicate DNA. The main DNA polymerases in prokaryotes are polymerases I, II, and III, while in eukaryotes they are polymerases α, δ, ε, and γ. RNA polymerases were discovered in 1960 and synthesize different RNA molecules. The three types of RNA polymerase in cells are polymerases I, II, and III, which produce rRNAs, mRNAs/hnRNAs, and tRNAs/rRNAs, respectively. Poly