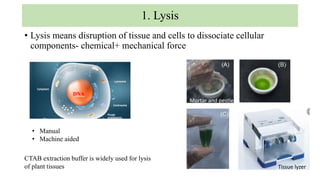
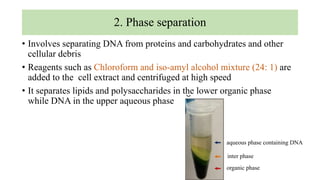
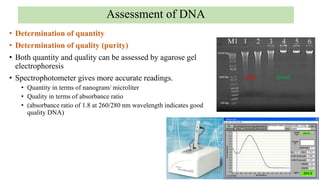
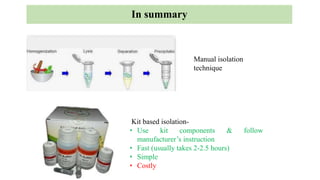
DNA can be isolated from cells using various techniques. The basic steps involve lysing the cells to release DNA, separating DNA from other biomolecules using chemicals like chloroform, precipitating purified DNA using ethanol, washing away impurities, and resuspending the DNA. Assessment of DNA quantity and quality is done using gel electrophoresis and spectrophotometry. Isolated DNA has many applications including genetic studies, identifying genes for important traits, and disease diagnosis.