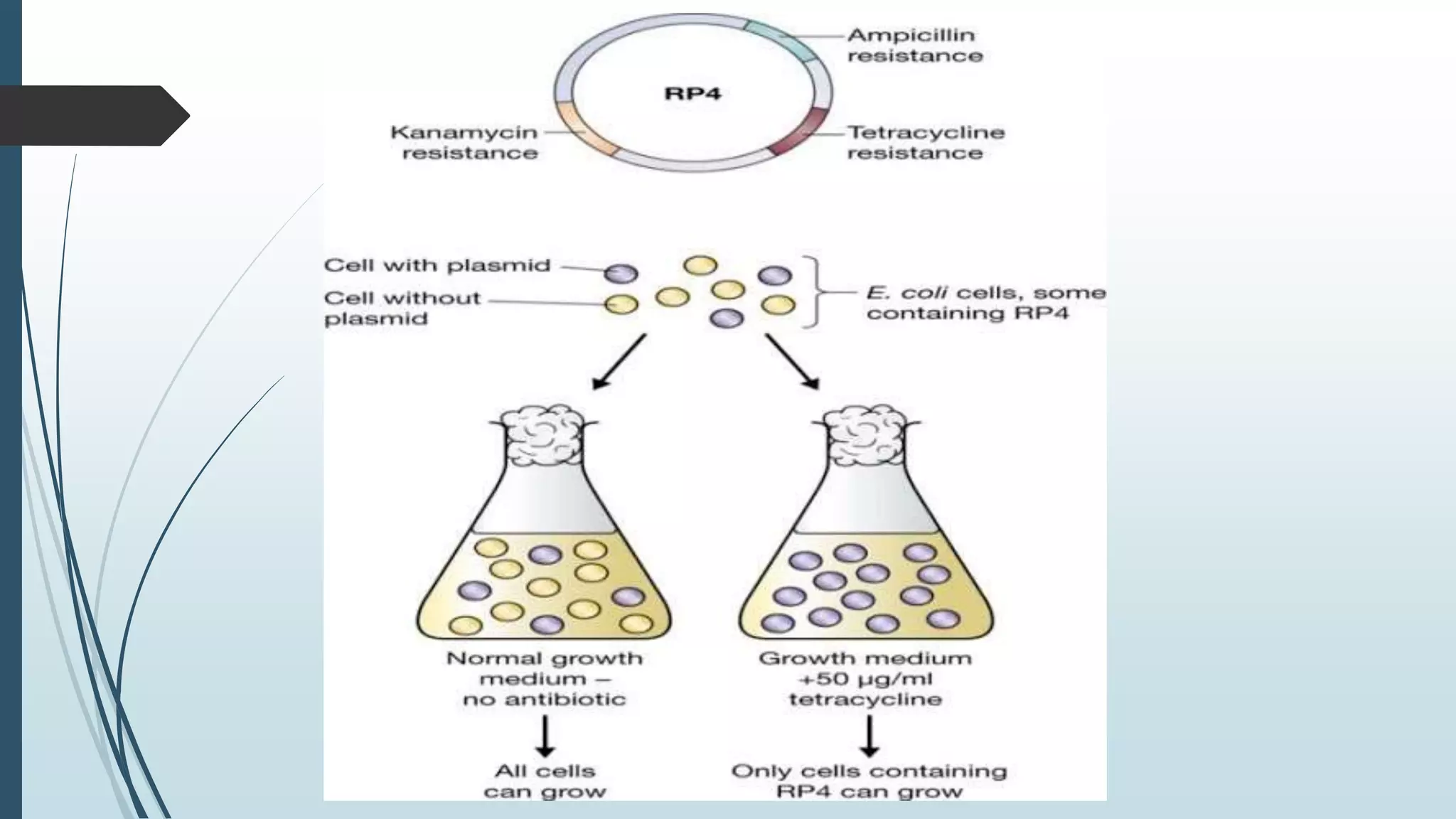
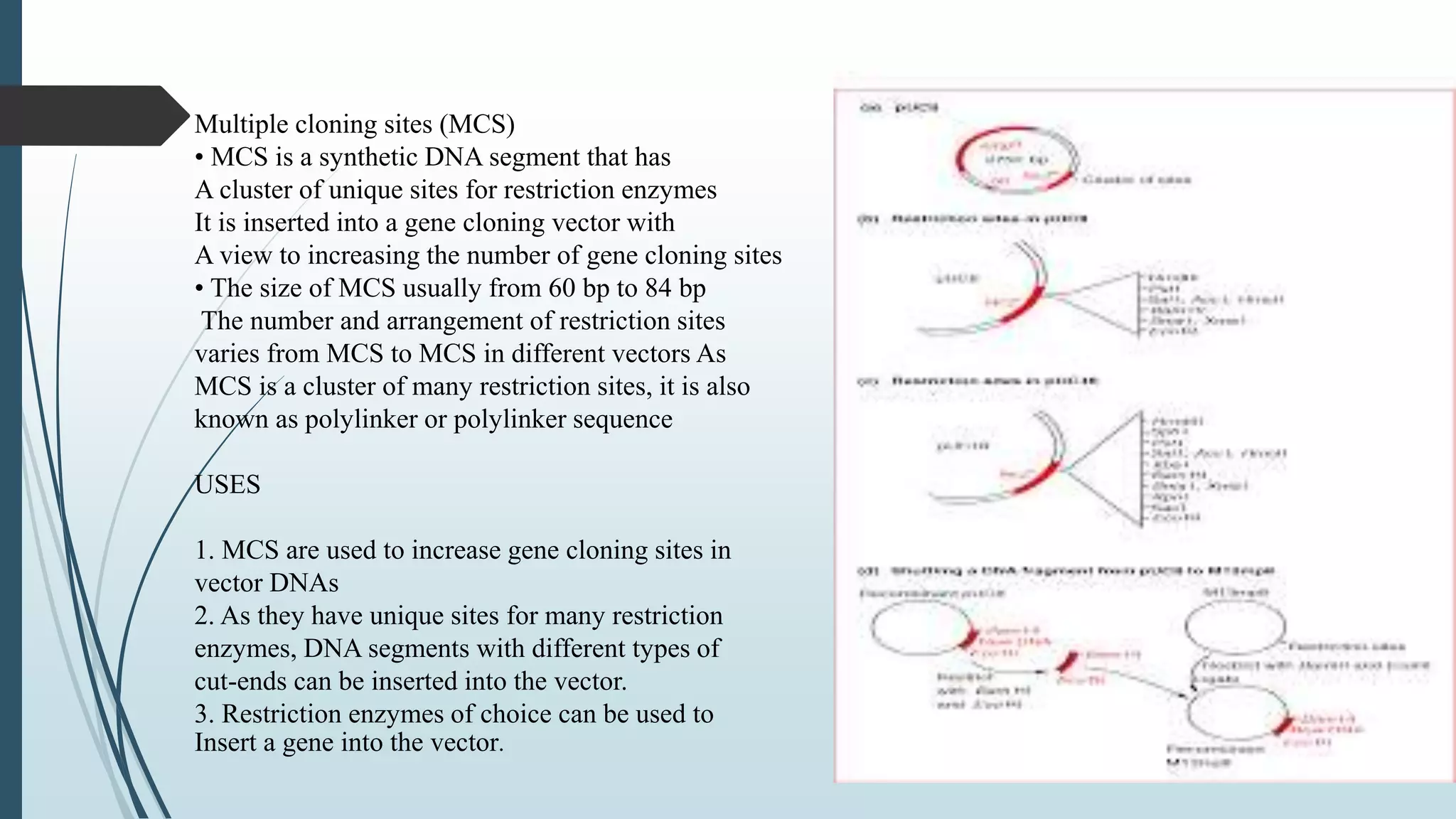
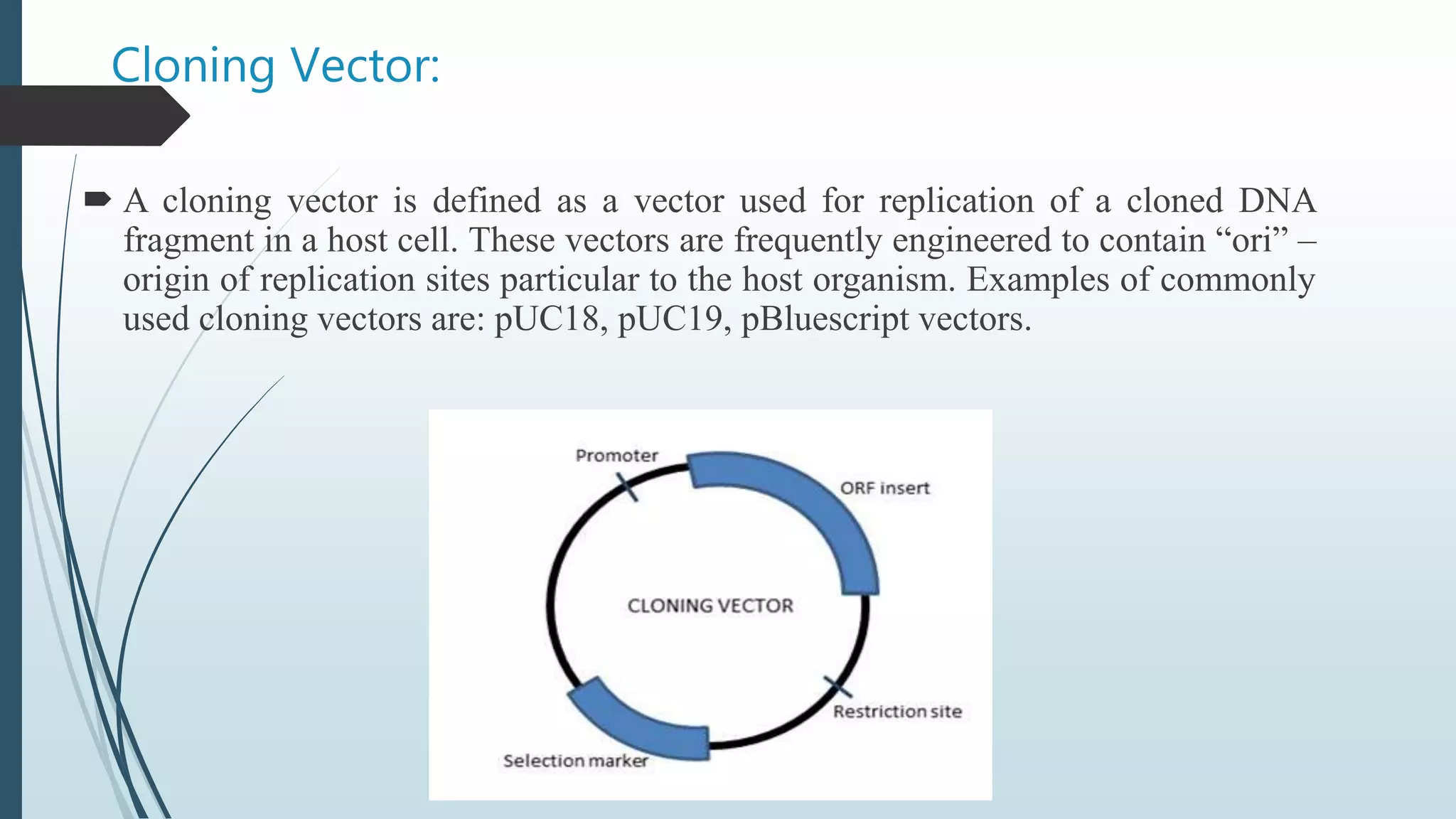
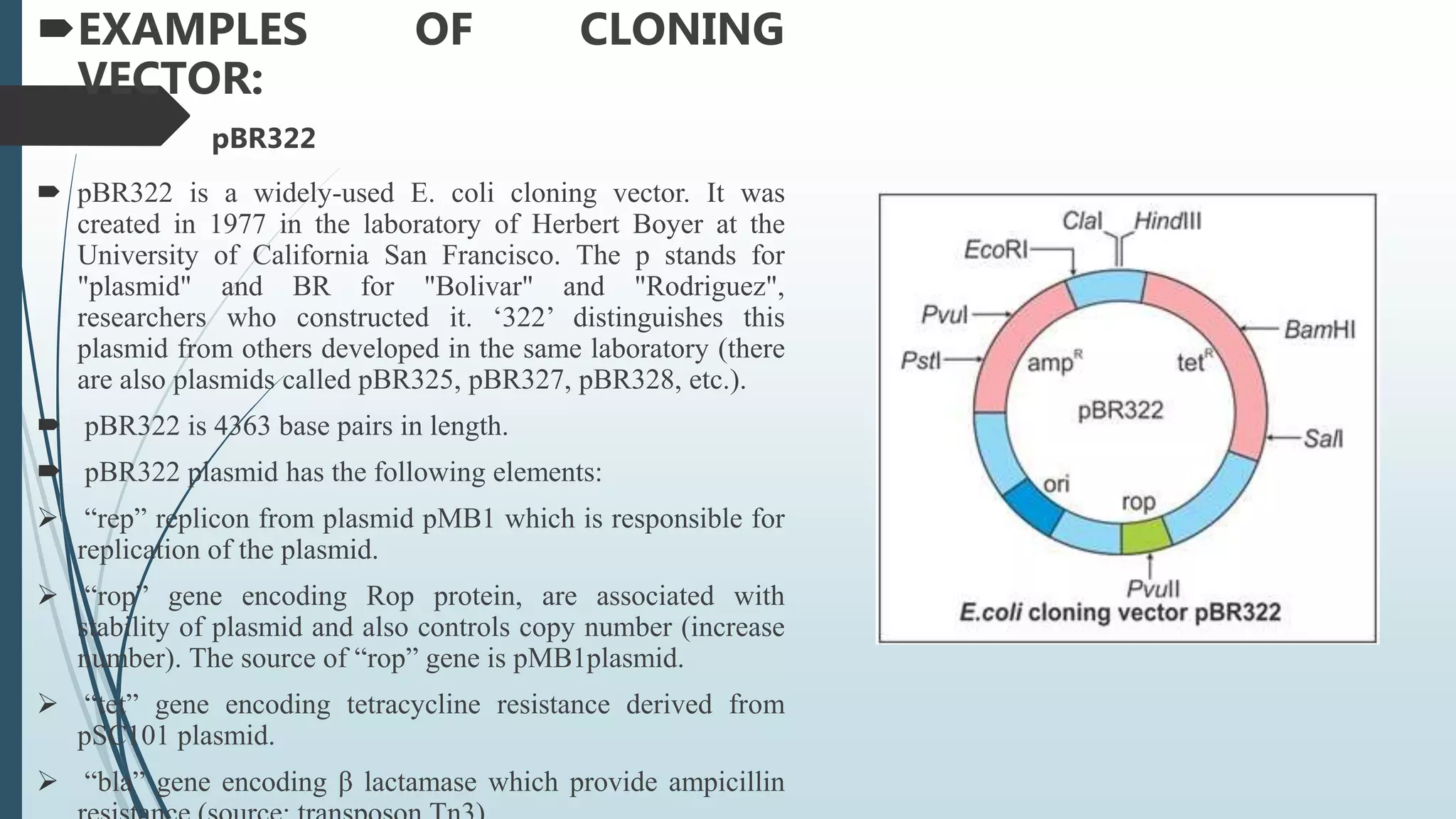
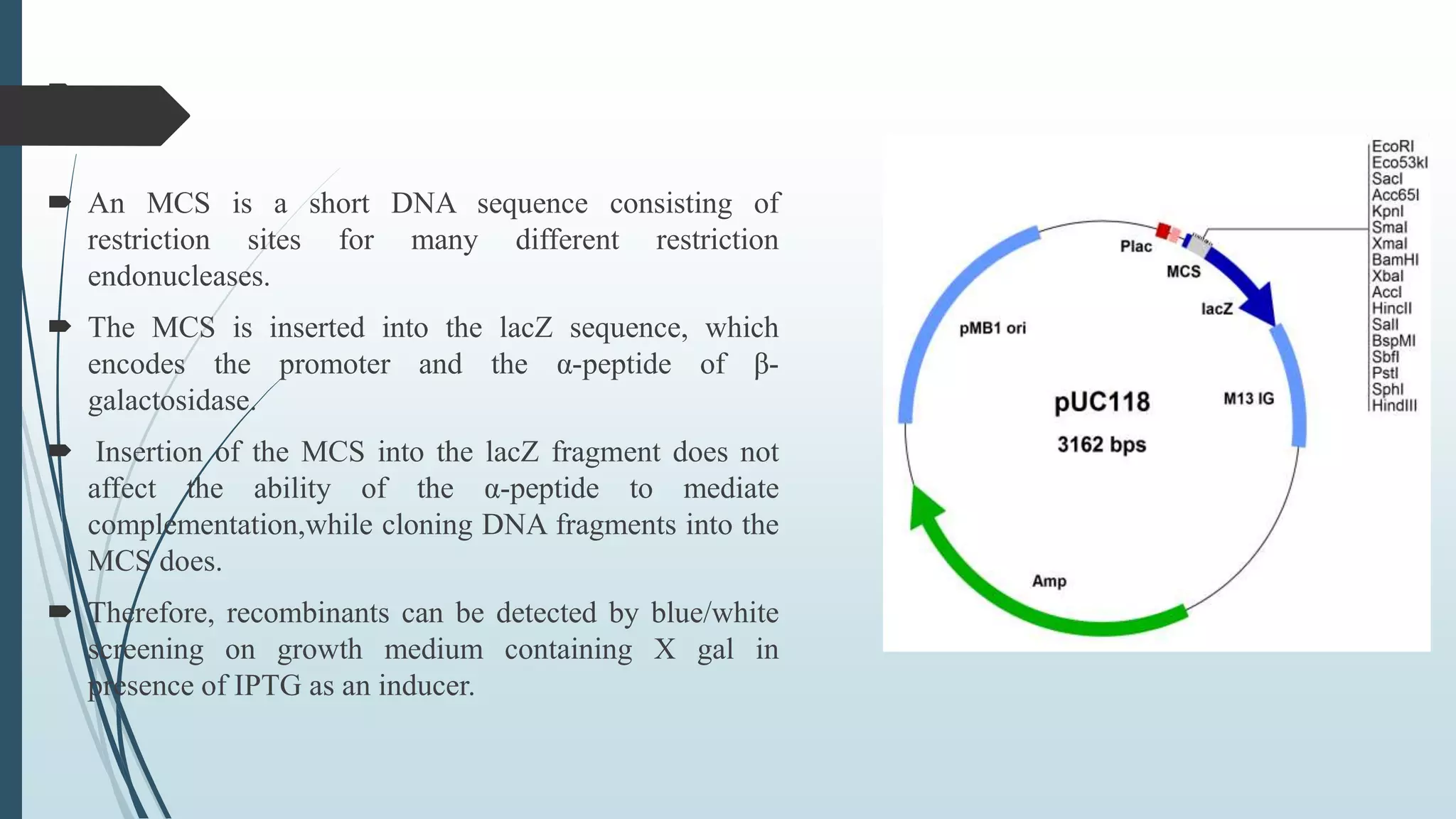
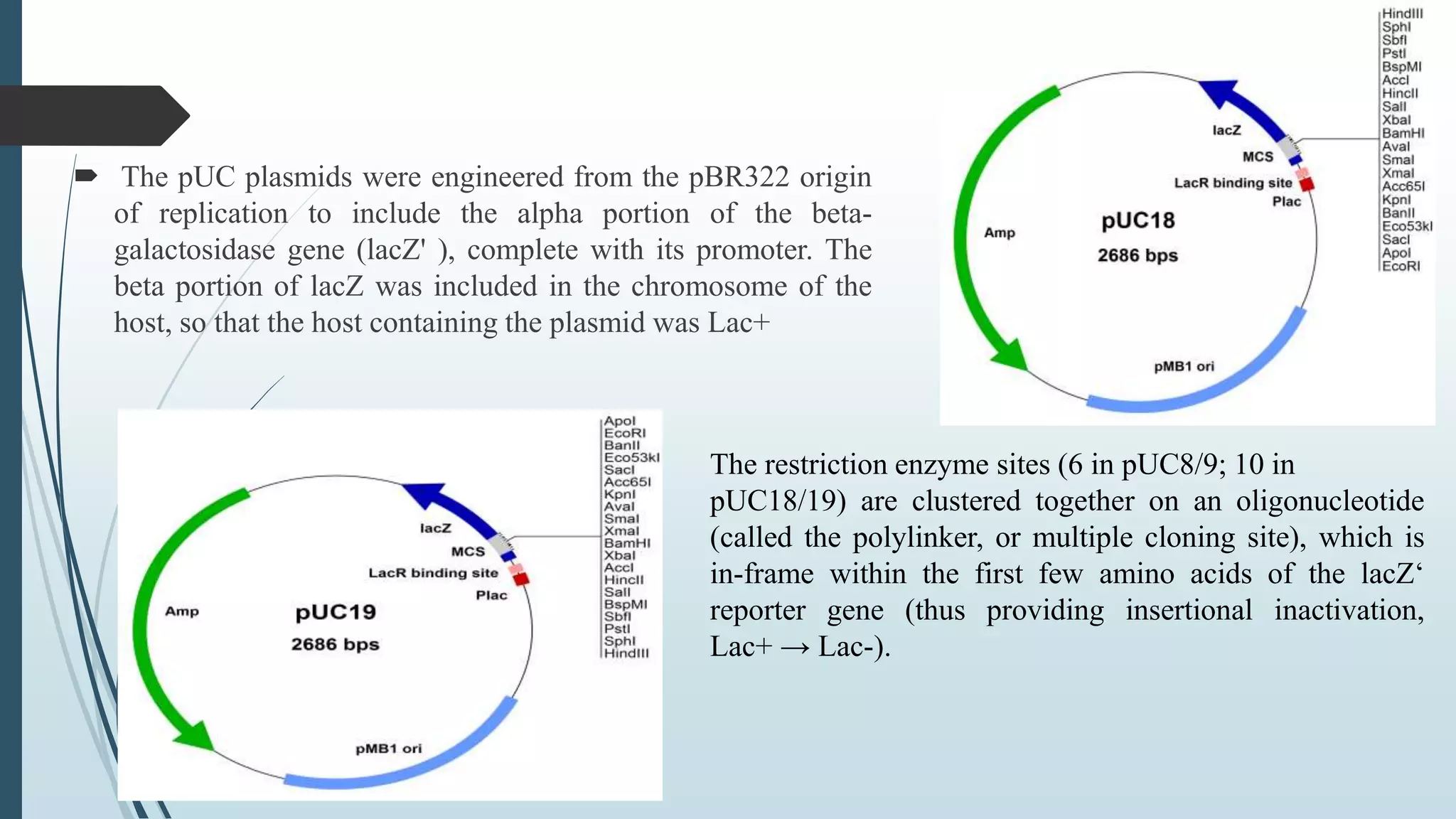
Plasmid vectors like pBR322 and pUC are commonly used cloning vectors. pBR322 was one of the first vectors created and has advantages like a small size, antibiotic resistance markers, and a high copy number. pUC vectors also have a small size and high copy number, and contain a multiple cloning site within the lacZ gene allowing visual selection of recombinants. Artificial vectors combine elements from different sources to overcome limitations of natural plasmids, and are designed for efficient cloning and expression of foreign DNA in host cells.