The caret package allows users to streamline the process of creating predictive models. It contains tools for data splitting, pre-processing, feature selection, model tuning using resampling, and variable importance estimation. The document provides examples of using various caret functions for visualization, pre-processing, model training and tuning, performance evaluation, and feature selection.



![A very simple example
library(caret)
str(iris)
set.seed(1)
#preprocess
process<-preProcess(iris[,-5],method=c('center','scale'))
dataScaled<-predict(process,iris[,-5])
#datasplitting
inTrain<-createDataPartition(iris$Species,p=0.75)[[1]]
length(inTrain)
trainData<-dataScaled[inTrain,]
trainClass<-iris[inTrain,5]
testData<-dataScaled[-inTrain,]
testClass<-iris[-inTrain,5]
/](https://image.slidesharecdn.com/dataminingwithcaretpackage-151214022117/85/Data-mining-with-caret-package-5-320.jpg)

![visualizations
The featurePlot function is a wrapper for different lattice plots to visualize the data.
Scatterplot Matrix
boxplot
featurePlot(x=iris[,1:4],
y=iris$Species,
plot="pairs",
##Addakeyatthetop
auto.key=list(columns=3))
featurePlot(x=iris[,1:4],
y=iris$Species,
plot="box",
##Addakeyatthetop
auto.key=list(columns=3))
/](https://image.slidesharecdn.com/dataminingwithcaretpackage-151214022117/85/Data-mining-with-caret-package-7-320.jpg)

![pre-processing
Zero- and Near Zero-Variance Predictors
data<-data.frame(x1=rnorm(100),
x2=runif(100),
x3=rep(c(0,1),times=c(2,98)),
x4=rep(3,length=100))
nzv<-nearZeroVar(data,saveMetrics=TRUE)
nzv
nzv<-nearZeroVar(data)
dataFilted<-data[,-nzv]
head(dataFilted)
/](https://image.slidesharecdn.com/dataminingwithcaretpackage-151214022117/85/Data-mining-with-caret-package-9-320.jpg)
![pre-processing
Identifying Correlated Predictors
set.seed(1)
x1<-rnorm(100)
x2<-x1+rnorm(100,0.1,0.1)
x3<-x1+rnorm(100,1,1)
data<-data.frame(x1,x2,x3)
corrmatrix<-cor(data)
highlyCor<-findCorrelation(corrmatrix,cutoff=0.75)
dataFilted<-data[,-highlyCor]
head(dataFilted)
/](https://image.slidesharecdn.com/dataminingwithcaretpackage-151214022117/85/Data-mining-with-caret-package-10-320.jpg)
![pre-processing
Identifying Linear Dependencies Predictors
set.seed(1)
x1<-rnorm(100)
x2<-x1+rnorm(100,0.1,0.1)
x3<-x1+rnorm(100,1,1)
x4<-x2+x3
data<-data.frame(x1,x2,x3,x4)
comboInfo<-findLinearCombos(data)
dataFilted<-data[,-comboInfo$remove]
head(dataFilted)
/](https://image.slidesharecdn.com/dataminingwithcaretpackage-151214022117/85/Data-mining-with-caret-package-11-320.jpg)

![pre-processing
Imputation:bagImpute/knnImpute/
data<-iris[,-5]
data[1,2]<-NA
data[2,1]<-NA
impu<-preProcess(data,method='knnImpute')
dataProced<-predict(impu,data)
/](https://image.slidesharecdn.com/dataminingwithcaretpackage-151214022117/85/Data-mining-with-caret-package-13-320.jpg)
![pre-processing
transformation: BoxCox/PCA
data<-iris[,-5]
pcaProc<-preProcess(data,method='pca')
dataProced<-predict(pcaProc,data)
head(dataProced)
/](https://image.slidesharecdn.com/dataminingwithcaretpackage-151214022117/85/Data-mining-with-caret-package-14-320.jpg)
![data splitting
create balanced splits of the data
set.seed(1)
trainIndex<-createDataPartition(iris$Species,p=0.8,list=FALSE, times=1)
head(trainIndex)
irisTrain<-iris[trainIndex,]
irisTest<-iris[-trainIndex,]
summary(irisTest$Species)
createResample can be used to make simple bootstrap samples
createFolds can be used to generate balanced cross–validation groupings from a set of data.
·
·
/](https://image.slidesharecdn.com/dataminingwithcaretpackage-151214022117/85/Data-mining-with-caret-package-15-320.jpg)

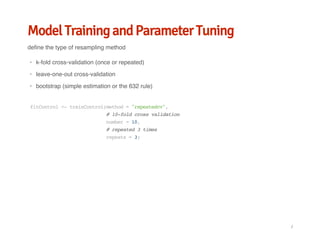
![Model Training and Parameter Tuning
prepare data
data(PimaIndiansDiabetes2,package='mlbench')
data<-PimaIndiansDiabetes2
library(caret)
#scaleandcenter
preProcValues<-preProcess(data[,-9],method=c("center","scale"))
scaleddata<-predict(preProcValues,data[,-9])
#YeoJohnsontransformation
preProcbox<-preProcess(scaleddata,method=c("YeoJohnson"))
boxdata<-predict(preProcbox,scaleddata)
/](https://image.slidesharecdn.com/dataminingwithcaretpackage-151214022117/85/Data-mining-with-caret-package-17-320.jpg)
![Model Training and Parameter Tuning
prepare data
#bagimpute
preProcimp<-preProcess(boxdata,method="bagImpute")
procdata<-predict(preProcimp,boxdata)
procdata$class<-data[,9]
#datasplitting
inTrain<-createDataPartition(procdata$class,p=0.75)[[1]]
length(inTrain)
trainData<-procdata[inTrain,1:8]
trainClass<-procdata[inTrain,9]
testData<-procdata[-inTrain,1:8]
testClass<-procdata[-inTrain,9]
/](https://image.slidesharecdn.com/dataminingwithcaretpackage-151214022117/85/Data-mining-with-caret-package-18-320.jpg)