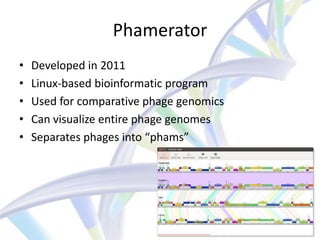
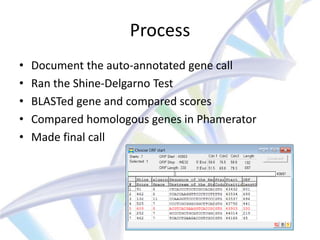
This document summarizes a CNIT final presentation about annotating the genome of the RiverMonster bacteriophage. It includes an introduction to bioinformatics and the goals of the CNIT course. It describes the bacteriophage and annotation tools used, including DNA Master, Phamerator, Glimmer, and GeneMark. It outlines the process of organizing the genome annotation between group members and analyzing each gene. It concludes with the accomplishments of completing the genome annotation and discusses the significance and potential future work.