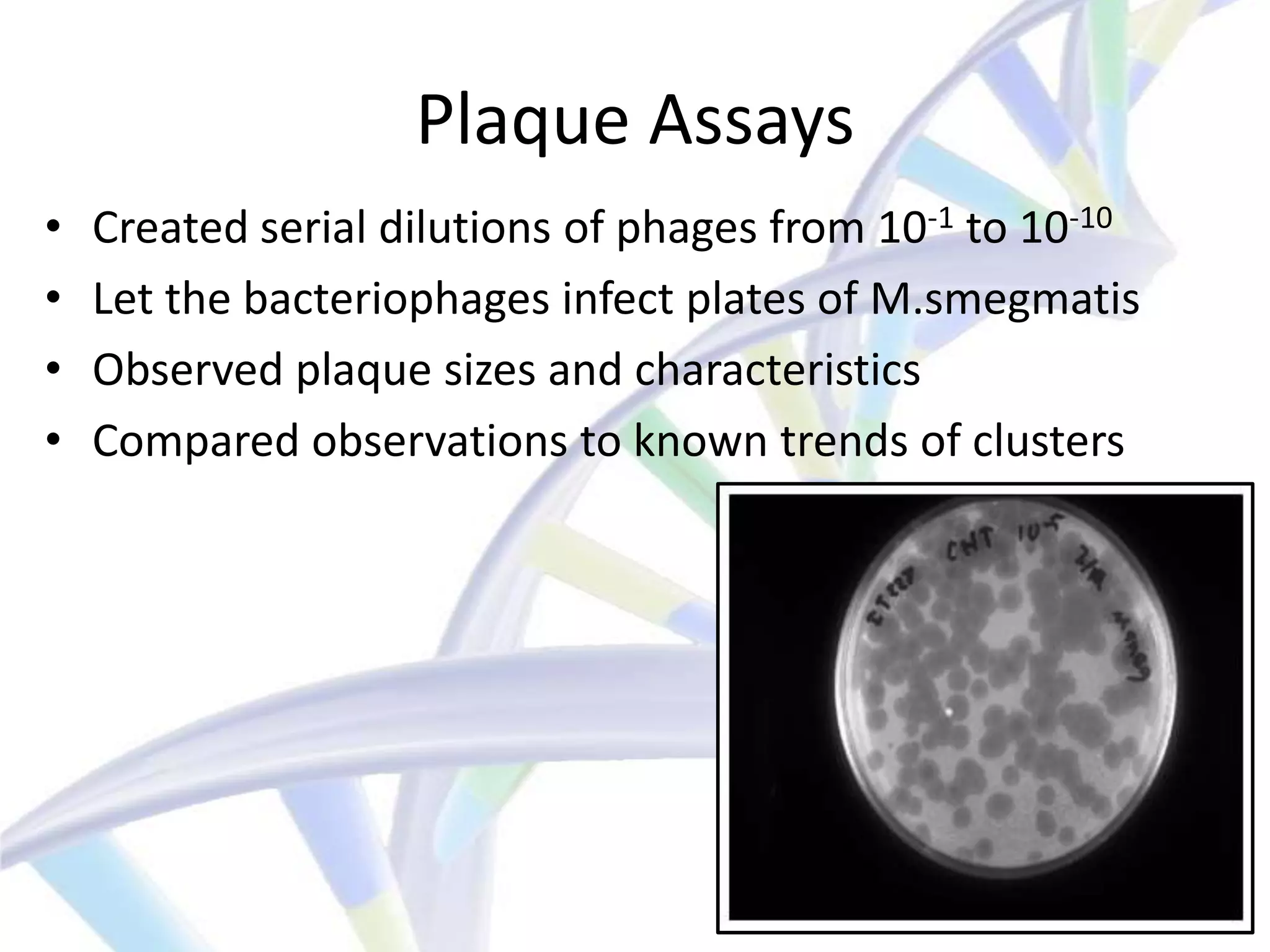
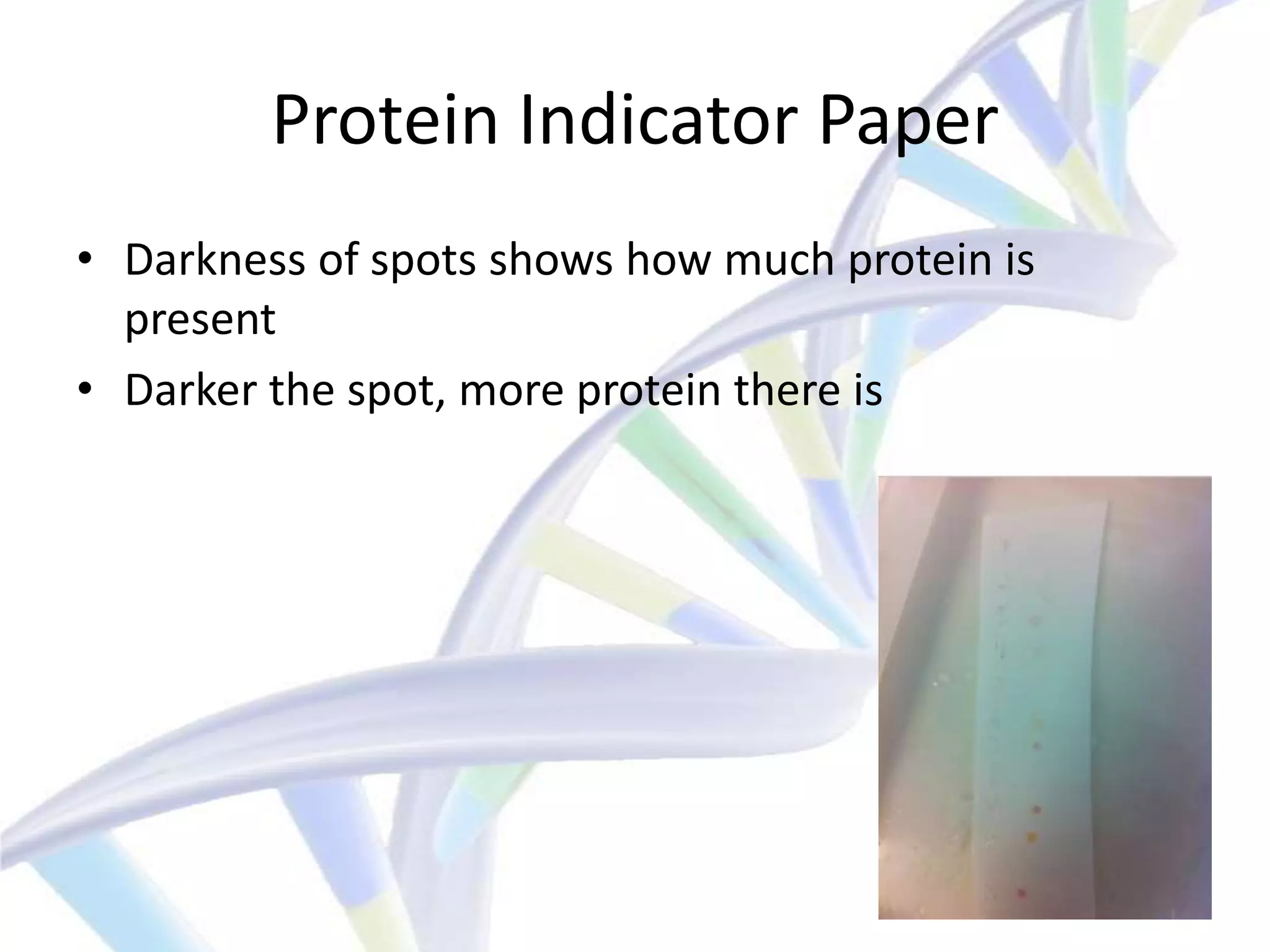
The document summarizes a student's IT final presentation project where they analyzed an unknown bacteriophage sample to determine its cluster assignment. The student performed experiments like plaque assays, DNA extraction, protein analysis, and mass spectrometry. By comparing the results to known cluster properties, the student concluded the phage belonged to cluster A, subcluster A3. The presentation discussed how this cluster analysis method could help schools without sequencing capabilities characterize phages.