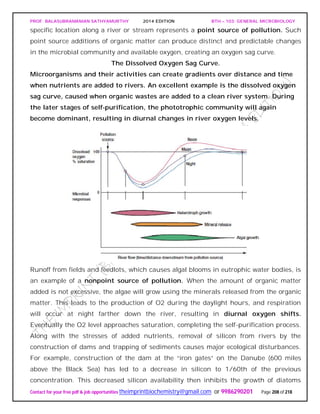
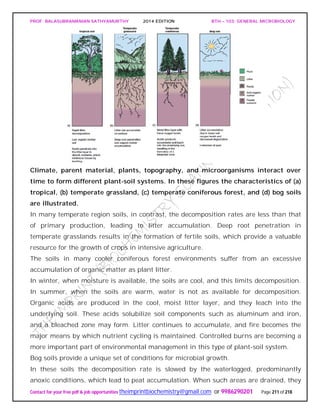
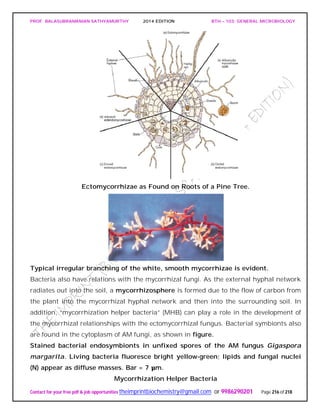
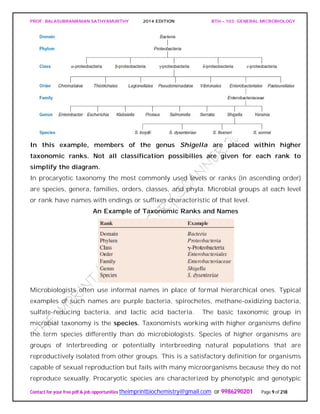
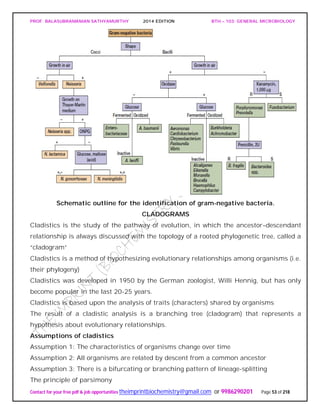
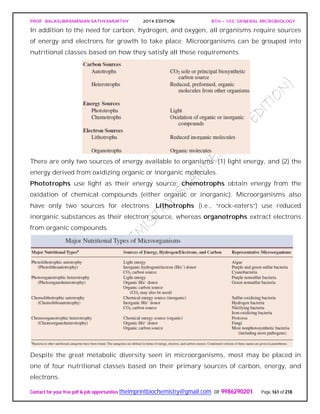
The document discusses microbial classification and taxonomy. It describes the three domain system of classification for microorganisms into Bacteria, Archaea, and Eukarya. It discusses phylogenetic relationships and the endosymbiotic theory of cellular evolution. It also outlines the code for bacterial nomenclature and taxonomy, including the hierarchical taxonomic ranks and criteria for classifying microbes, such as morphological, biochemical, and nucleic acid-based methods.












![PROF. BALASUBRAMANIAN SATHYAMURTHY 2014 EDITION BTH – 103: GENERAL MICROBIOLOGY
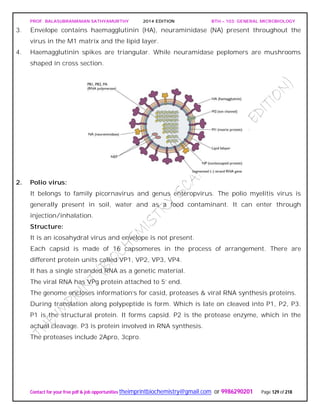
Contact for your free pdf & job opportunities theimprintbiochemistry@gmail.com or 9986290201 Page 15 of 218
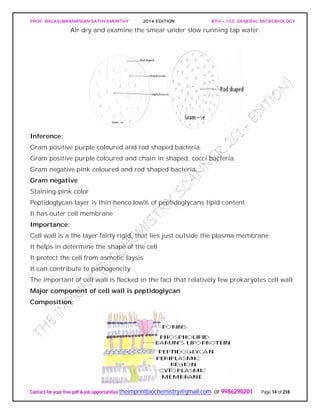
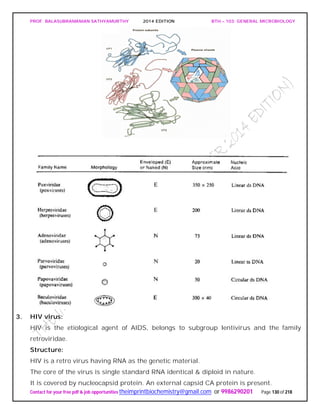
PEPTIDOGLYCAN substance present in most gram positive and gram negative
organism.
It has two parts
Glycan: It is made up of N-acetyl glucosomine and N-acetyl muramic acid linked by β-
1,4 glycosidic linkage.
NAG.............NAM [Lactyl ester of NAG]
Peptide: It consists of two parts:
Tetra peptide: It consists of various D and L form of amino acid connected to carboxyl
group of N-acetyl mumaric acid. They are:
Gram negative - L-alanine,D-glutamic acid,diamino pimelic acid,D-alanine.
Peptide linkage:
Peptide linkage is formed between D-alanine and L-lysine (in case of gram +ve) and
diamino primalic acid (in case of gram –ve)
The peptide chain (containing &amino acids) are linked to “NAM” and a peptide chain in
the two side chains.
In gram negative cell wall inter chain peptide linkage is a CO-NH linkage
In the lactyl group because of the free C=0 group (in NAM peptide) chain is attached to
it for example in gram negative cell wall
Peptidoglycan substance present in most gram positive and gram negative organism.
It has two parts
Glycan: It is made up of N-acetyl glucosomine and N-acetyl muramic acid linked by β-
1, 4 glycosidic linkages.
NAG.............NAM [Lactyl ester of NAG]
Peptide: It consists of two parts:
Tetra peptide: It consists of various D and L form of amino acid connected to carboxyl
group of N-acetyl mumaric acid. They are:-
Gram negative - L-alanine, D-glutamic acid, diamino pimelic acid,D-alanine.
Peptide linkage:
Peptide linkage is formed between D-alanine and L-lysine (in case of gram +ve) and
diamino primalic acid (in case of gram –ve)
The peptide chain (containing &amino acids) are linked to “NAM” and a peptide chain in
the two side chains.
In gram negative cell wall inter chain peptide linkage is a CO-NH linkage](https://image.slidesharecdn.com/bth103generalmicrobiology-190421182551/85/Bth-103-general-microbiology-15-320.jpg)















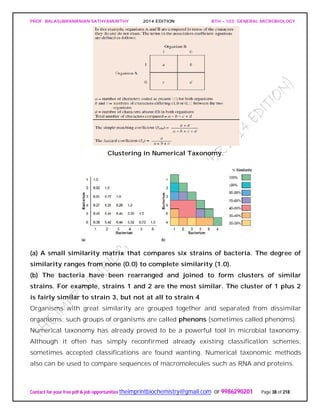





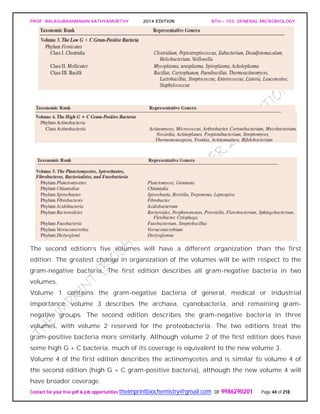
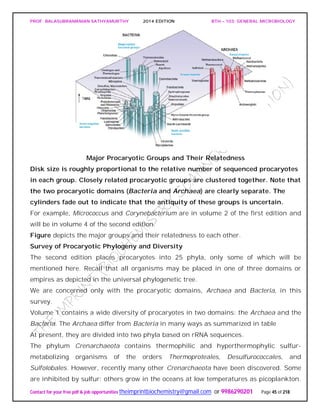






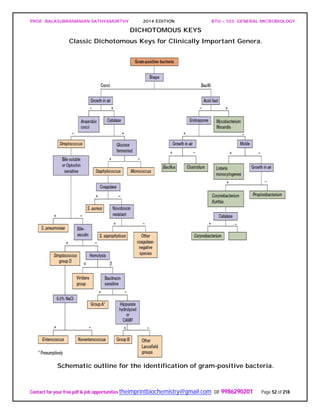
![PROF. BALASUBRAMANIAN SATHYAMURTHY 2014 EDITION BTH – 103: GENERAL MICROBIOLOGY
Contact for your free pdf & job opportunities theimprintbiochemistry@gmail.com or 9986290201 Page 56 of 218
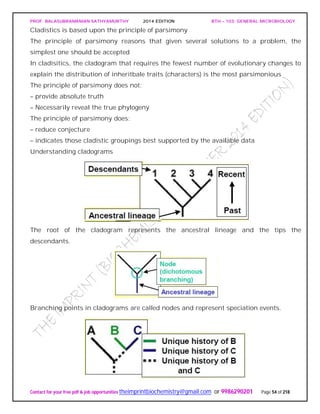
appear to be one. The ordinate in such a dendrogram has no special significance, and
the clusters may be arranged in any convenient order.
The significance of these clusters or phenons in traditional taxonomic terms is not
always evident, and the similarity levels at which clusters are labeled species, genera,
and so on, are a matter of judgment. Sometimes groups are simply called phenons and
preceded by a number showing the similarity level above which they appear (e.g., a 70-
phenon is a phenon with 70% or greater similarity among its constituents). Phenons
formed at about 80% similarity often are equivalent to species.
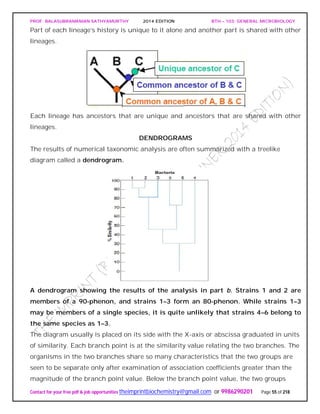
UNIVERSAL PHYLOGENETIC TREE
Following the publication in 1859 of Darwin’s On the Origin of Species, biologists began
trying to develop phylogenetic or phyletic classification systems. These are systems
based on evolutionary relationships rather than general resemblance (the term
phylogeny [Greek phylon, tribe or race, and genesis, generation or origin] refers to the
evolutionary development of a species). This has proven difficult for procaryotes and
other microorganisms, primarily because of the lack of a good fossil record. The direct
comparison of genetic material and gene products such as RNA and proteins overcomes
many of these problems.
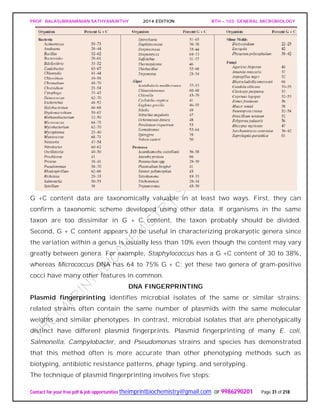
The studies of Carl Woese and his collaborators on rRNA sequences in procaryotic cells
suggest that procaryotes divided into two distinct groups very early on.
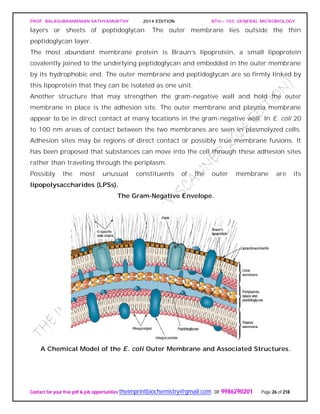
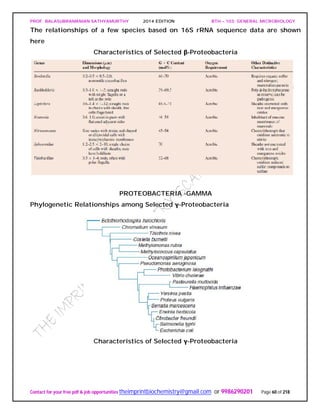
Figure depicts a universal phylogenetic tree that reflects these views. The tree is divided
into three major branches representing the three primary groups: Bacteria, Archaea,
and Eucarya. The archaea and bacteria first diverged, and then the eucaryotes
developed. These three primary groups are called domains and placed above the
phylum and kingdom levels (the traditional kingdoms are distributed among these three
domains). The domains differ markedly from one another.
Eucaryotic organisms with primarily glycerol fatty acyl diester membrane lipids and
eucaryotic rRNA belong to the Eucarya. The domain Bacteria contains procaryotic
cells with bacterial rRNA and membrane lipids that are primarily diacyl glycerol
diesters. Procaryotes having isoprenoid glycerol diether or diglycerol tetraether lipids in
their membranes and archaeal rRNA compose the third domain, Archaea.
It appears likely that modern eucaryotic cells arose from procaryotes about 1.4 billion
years ago. There has been considerable speculation about how eucaryotes might have](https://image.slidesharecdn.com/bth103generalmicrobiology-190421182551/85/Bth-103-general-microbiology-56-320.jpg)


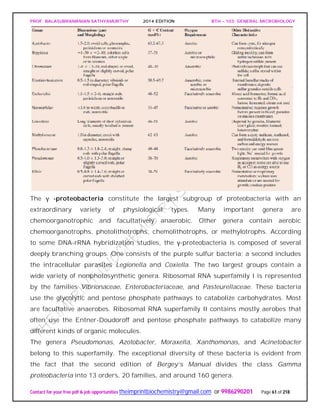
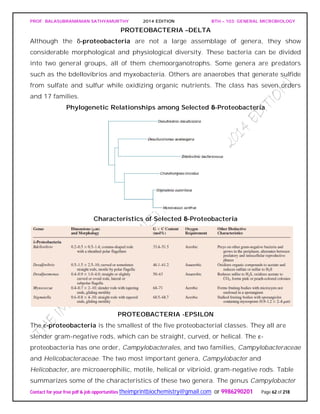









![PROF. BALASUBRAMANIAN SATHYAMURTHY 2014 EDITION BTH – 103: GENERAL MICROBIOLOGY
Contact for your free pdf & job opportunities theimprintbiochemistry@gmail.com or 9986290201 Page 75 of 218
ACTINOBACTERIA
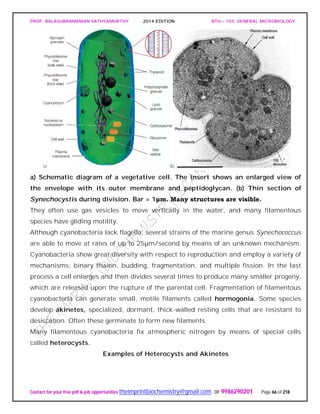
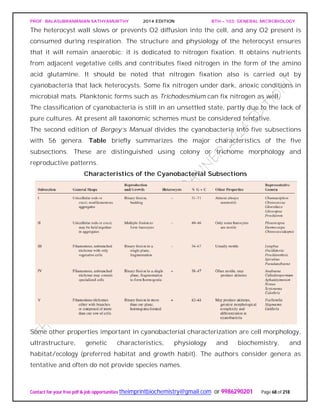
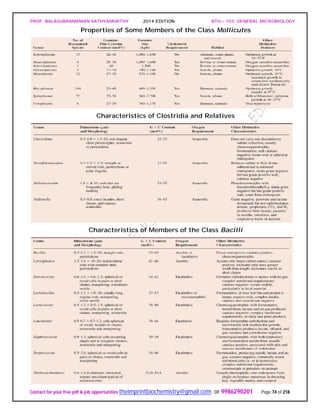
General Properties of the Actinomycetes
When growing on a solid substratum such as agar, the branching network of hyphae
developed by actinomycetes grows both on the surface of the substratum and into it to
form a substrate mycelium. Septa usually divide the hyphae into long cells (20 μm and
longer) containing several nucleoids. Sometimes a tissuelike mass results and may be
called a thallus. Many actinomycetes also have an aerial mycelium that extends above
the substratum and forms asexual, thin-walled spores called conidia [s., conidium] or
conidiospores on the ends of filaments.
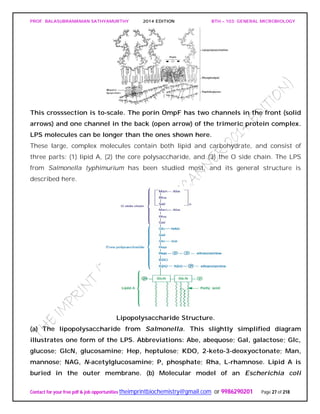
An Actinomycete Colony.
The cross section of an actinomycete colony with living (blue-green) and dead
(white) hyphae. The substrate mycelium and aerial mycelium with chains of
conidiospores are shown.
If the spores are located in a sporangium, they are called sporangiospores. The spores
can vary greatly in shape.
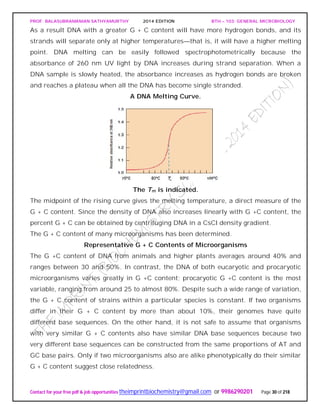
Examples of Actinomycete Spores as Seen in the Scanning Electron Microscope.
Sporulating Saccharopolyspora hyphae (X3,000).](https://image.slidesharecdn.com/bth103generalmicrobiology-190421182551/85/Bth-103-general-microbiology-75-320.jpg)
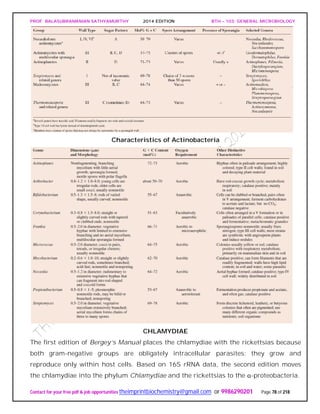

![PROF. BALASUBRAMANIAN SATHYAMURTHY 2014 EDITION BTH – 103: GENERAL MICROBIOLOGY
Contact for your free pdf & job opportunities theimprintbiochemistry@gmail.com or 9986290201 Page 81 of 218
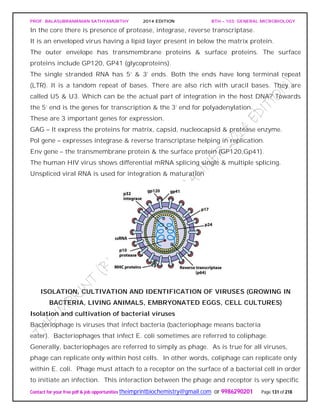
that acquires host ATP in exchange for ADP. Thus chlamydiae seem to be energy
parasites that are completely dependent on their hosts for ATP. However, this might not
be the complete story. The complete sequence of the C. trachomatis genome indicates
that the bacterium may be able to synthesize at least some ATP. Although there are two
genes for ATP/ADP translocases, there also are genes for substrate-level
phosphorylation, electron transport, and oxidative phosphorylation.
When supplied with precursors from the host, RBs can synthesize DNA, RNA, glycogen,
lipids, and proteins. Presumably the RBs have porins and active membrane transport
proteins, but little is known about these. They also can synthesize at least some amino
acids and coenzymes. The EBs has minimal metabolic activity and cannot take in ATP
or synthesize proteins. They seem to be dormant forms concerned exclusively with
transmission and infection.
Three chlamydial species are important pathogens of humans and other warm-blooded
animals. C. trachomatis infects humans and mice. In humans it causes trachoma,
nongonococcal urethritis, and other diseases. C. psittaci causes psittacosis in humans.
However, unlike C. trachomatis, it also infects many other animals (e.g., parrots,
turkeys, sheep, cattle, and cats) and invades the intestinal, respiratory, and genital
tracts; the placenta and fetus; the eye; and the synovial fluid of joints.
Chlamydia pneumoniae is a common cause of human pneumonia. There is now indirect
evidence that infections by C. pneumonia may be associated with the development of
atherosclerosis and that chlamydial infections may cause severe heart inflammation
and damage. Recently a fourth species, C. pecorum, has been recognized.
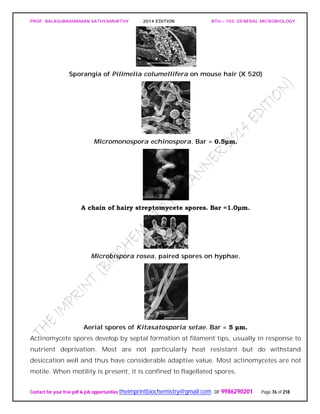
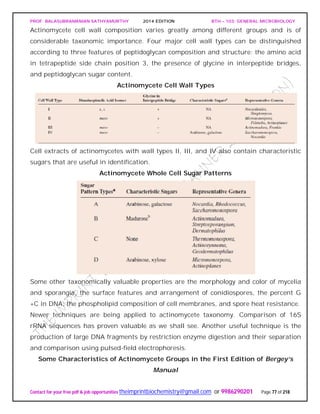
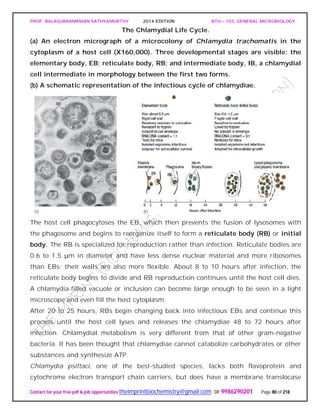
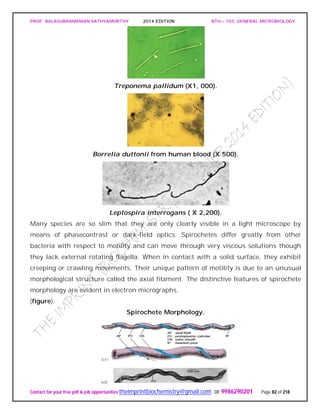
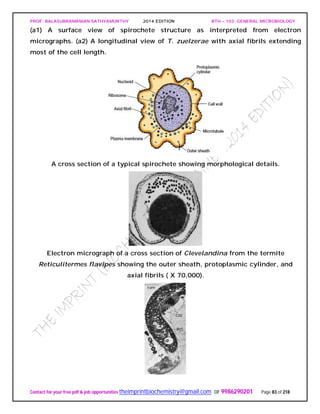
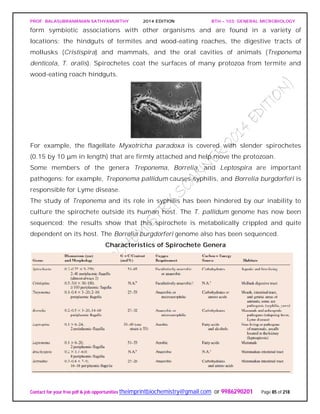
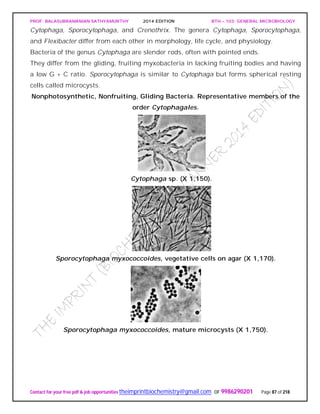
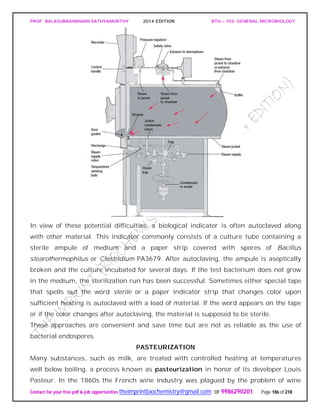
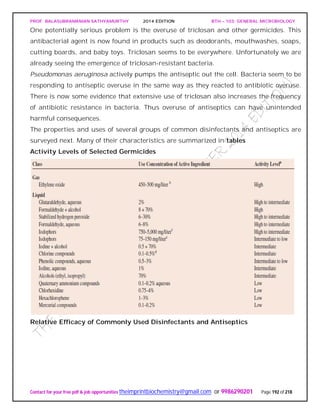
SPIROCHAETES
The phylum Spirochaetes [Greek spira, a coil, and chaete, hair] contains gram-negative,
chemoheterotrophic bacteria distinguished by their structure and mechanism of
motility. They are slender, long bacteria (0.1 to 3.0 μm by 5 to 250 μm) with a flexible,
helical shape.
The Spirochetes - Representative examples.
Cristispira sp. from the crystalline style of a clam; phase contrast (X2,200).](https://image.slidesharecdn.com/bth103generalmicrobiology-190421182551/85/Bth-103-general-microbiology-81-320.jpg)




![PROF. BALASUBRAMANIAN SATHYAMURTHY 2014 EDITION BTH – 103: GENERAL MICROBIOLOGY
Contact for your free pdf & job opportunities theimprintbiochemistry@gmail.com or 9986290201 Page 90 of 218
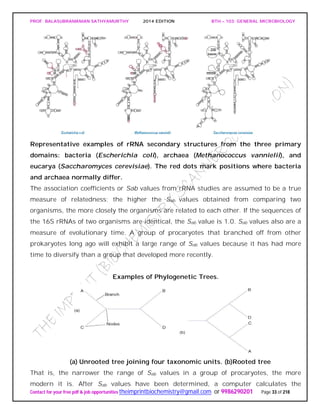
The first edition of Bergey’s Manual divided the archaea into five major groups based on
physiological and morphological differences.
Table summarizes some the characteristics of these five groups and gives
representatives of each.
Characteristics of the Major Archaeal Groups
On the basis of rRNA data, the second edition of Bergey’s Manual will divide the
archaea into the phyla Euryarchaeota [Greek eurus, wide, and Greek archaios, ancient
or primitive] and Crenarchaeota [Greek crene, spring or fount, and archaios]. The
euryarchaeotes are given this name because they occupy many different ecological
niches and have a variety of metabolic patterns.
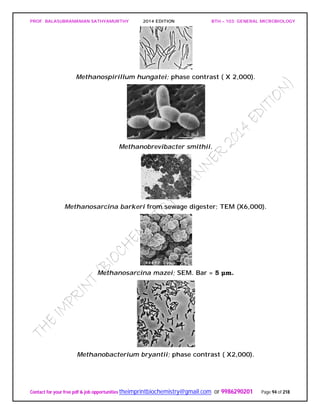
The phylum Euryarchaeota is very diverse with seven classes (Methanobacteria,
Methanococci, Halobacteria, Thermoplasmata, Thermococci, Archaeglobi, and
Methanopyri), nine orders, and 15 families. The methanogens, extreme halophiles,
sulfate reducers, and many extreme thermophiles with sulfur-dependent metabolism
are located in the Euryarchaeota. Methanogens are the dominant physiological group.
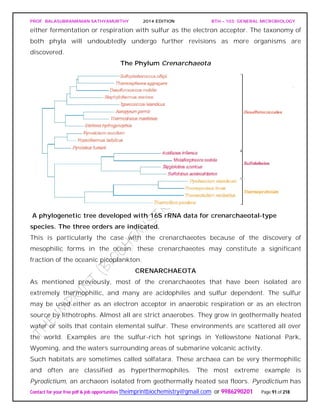
The crenarchaeotes are thought to resemble the ancestor of the archaea, and almost all
the well-characterized species are thermophiles or hyperthermophiles. The phylum
Crenarchaeota is divided into one class, Thermoprotei, and three orders. The order
Thermoproteales contains gram-negative anaerobic to facultative, hyperthermophilic
rods. They often grow chemolithoautotrophically by reducing sulfur to hydrogen sulfide.
Members of the order Sulfolobales are coccus-shaped thermoacidophiles. The order
Desulfurococcales contains gramnegative coccoid or disk-shaped hyperthermophiles.
They grow either chemolithotrophically by hydrogen oxidation or organotrophically by](https://image.slidesharecdn.com/bth103generalmicrobiology-190421182551/85/Bth-103-general-microbiology-90-320.jpg)



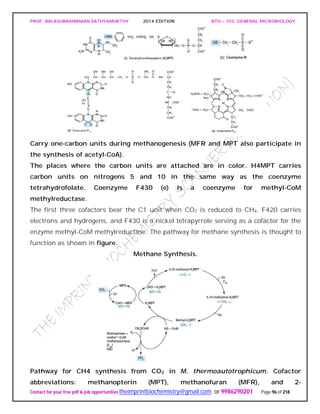



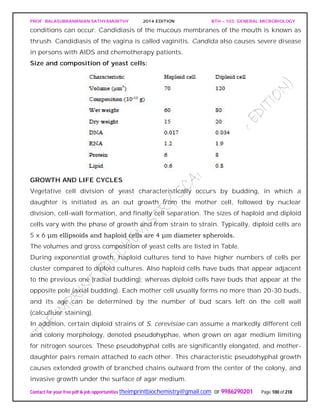
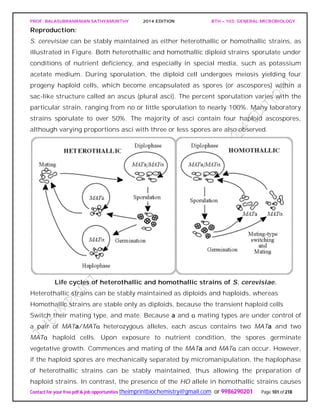



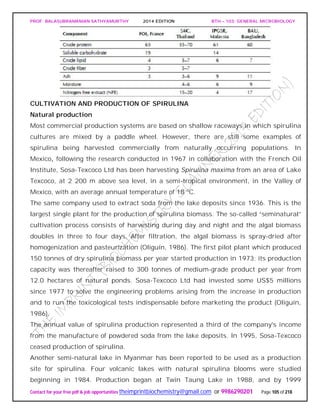

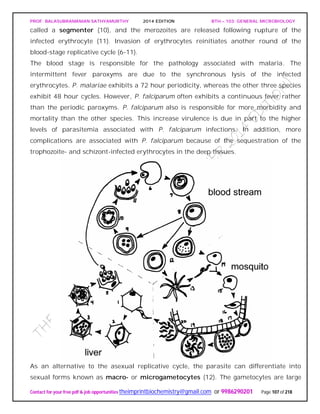

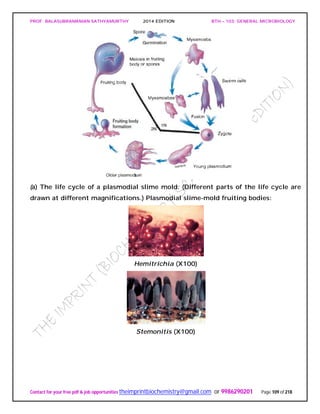

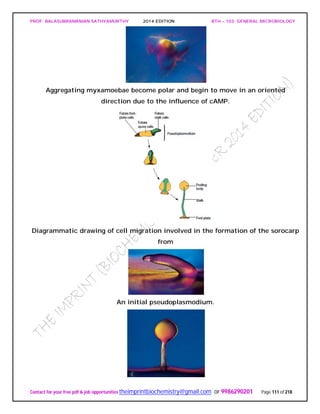






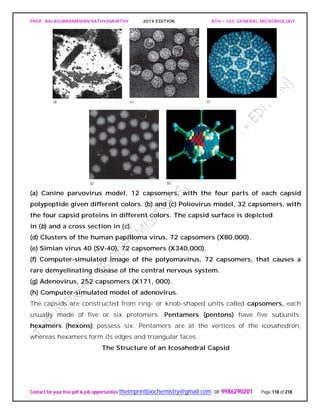
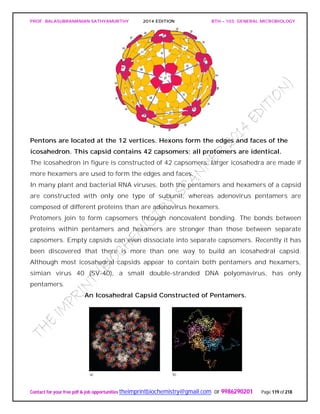



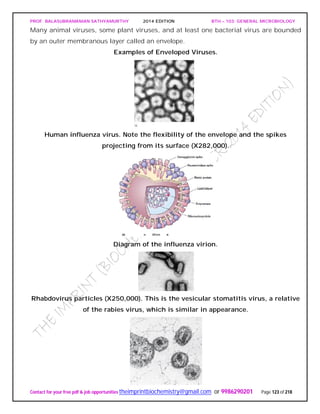
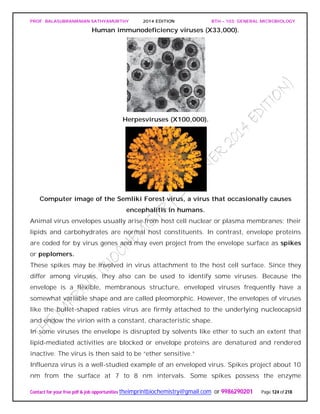

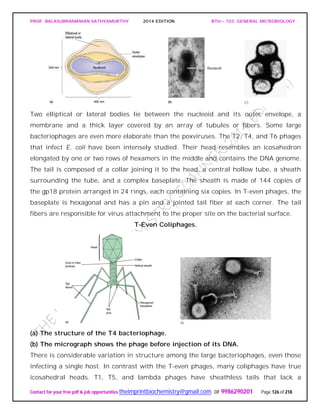




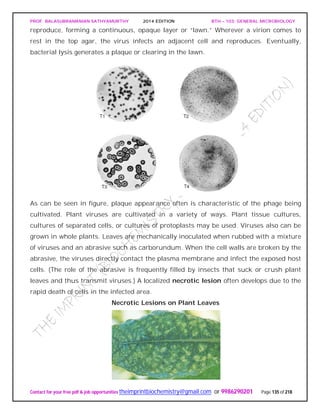


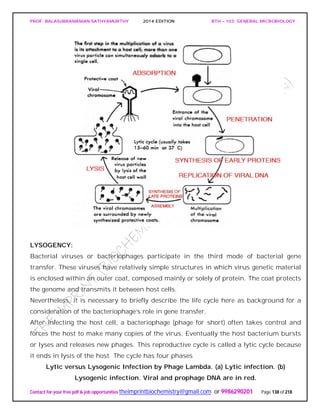
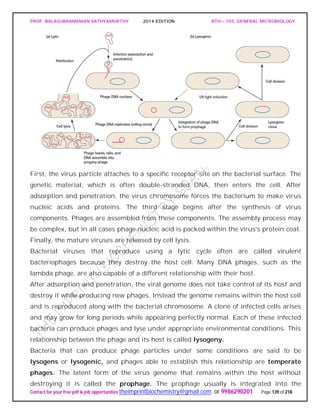
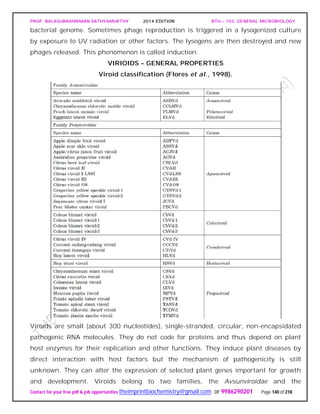

![PROF. BALASUBRAMANIAN SATHYAMURTHY 2014 EDITION BTH – 103: GENERAL MICROBIOLOGY
Contact for your free pdf & job opportunities theimprintbiochemistry@gmail.com or 9986290201 Page 142 of 218
invertebrates, and the prion protein is bound to the surface of neurons. Presumably an
altered PrP is at least partly responsible for the disease.
Despite the isolation of PrP, the mechanism of prion diseases continues to stir
controversy. Most researchers are convinced that prion diseases are transmitted by the
PrP alone. They believe that the infective pathogen is an abnormal PrP (PrPSc; Sc,
scrapieassociated), a protein that has either been changed in conformation or
chemically modified. When PrPSc enters a normal brain, it might bind to the normal
cellular PrP (PrPC) and induce it to fold into the abnormal conformation. The newly
produced PrPSc mo ecule could then convert more normal PrPC proteins to the PrPSc
form. Alternatively, PrPSc could activate enzymes that modify PrP structure. Prions with
different amino acid sequences and conformations convert PrpC molecules to PrPSc in
other hosts.
More support for the “protein-only” hypothesis has been supplied by studies on the
yeast protein Sup35, which aids in the termination of protein synthesis in
Saccharomyces cerevisiae. Because Sup35 acts much like mammalian prions, it has
been called a yeast prion. Sup35 exists in an inactive form in [PSI_] cells, and the
phenotype can be passed on to daughter cells. The inactive, insoluble
form of Sup35 aggregates and translation does not terminate properly. There is
evidence that the [PSI_] phenotype proliferates in yeast when the inactive prion form of
Sup35 interacts with normal, soluble Sup35 and induces a self-propagating
conformational change of active Sup35 to the inactive form.
A minority think that the “protein-only” hypothesis is inadequate. They are concerned
about the existence of prion strains, which they believe are genetic, and about the
problem of how genetic information can be transmitted between hosts by a protein.
Doubters note that proteins have never been known to carry genetic information. It
could be that prions somehow either directly aid an infectious agent such as an
unknown virus or increase susceptibility to the agent. Possibly the real agent is a tiny
nucleic acid that is coated with PrP and interacts with host cells to cause disease. This
hypothesis is consistent with the finding that many strains of the scrapie agent have
been isolated. There is also some evidence that a strain can change or mutate.
Supporters of the “protein-only” hypothesis reply that strain characteristics are simply
due to structural differences in the PrP molecule.](https://image.slidesharecdn.com/bth103generalmicrobiology-190421182551/85/Bth-103-general-microbiology-142-320.jpg)




![PROF. BALASUBRAMANIAN SATHYAMURTHY 2014 EDITION BTH – 103: GENERAL MICROBIOLOGY
Contact for your free pdf & job opportunities theimprintbiochemistry@gmail.com or 9986290201 Page 148 of 218
water activity or osmotic concentration. For example, Staphylococcus aureus can be
cultured in media containing any sodium chloride concentration up to about 3 M. It is
well adapted for growth on the skin. The yeast Saccharomyces rouxii will grow in sugar
solutions with aw values as low as 0.6. The alga Dunaliella viridis tolerates sodium
chloride concentrations from 1.7 M to a saturated solution. Although a few
microorganisms are truly osmotolerant, most only grow well at water activities around
0.98 (the approximate aw for seawater) or higher. This is why drying food or adding
large quantities of salt and sugar is so effective in preventing food spoilage.
Halophiles have adapted so completely to hypertonic, saline conditions that they
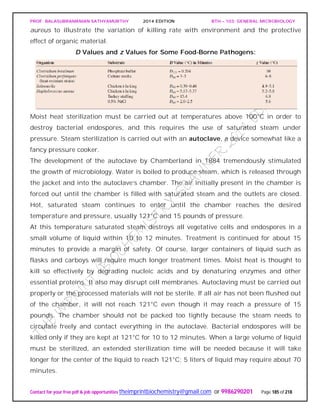
require high levels of sodium chloride to grow, concentrations between about 2.8 M and
saturation (about 6.2 M) for extreme halophilic bacteria. The archaeon Halobacterium
can be isolated from the Dead Sea (a salt lake between Israel and Jordan and the lowest
lake in the world), the Great Salt Lake in Utah, and other aquatic habitats with salt
concentrations approaching saturation. Halobacterium and other extremely halophilic
bacteria have significantly modified the structure of their proteins and membranes
rather than simply increasing the intracellular concentrations of solutes, the approach
used by most osmotolerant microorganisms. These extreme halophiles accumulate
enormous quantities of potassium in order to remain hypertonic to their environment;
the internal potassium concentration may reach 4 to 7 M. The enzymes, ribosomes, and
transport proteins of these bacteria require high levels of potassium for stability and
activity. In addition, the plasma membrane and cell wall of Halobacterium are stabilized
by high concentrations of sodium ion. If the sodium concentration decreases too much,
the wall and plasma membrane literally disintegrate. Extreme halophilic bacteria have
successfully adapted to environmental conditions that would destroy most organisms.
In the process they have become so specialized that they have lost ecological flexibility
and can prosper only in a few extreme habitats.
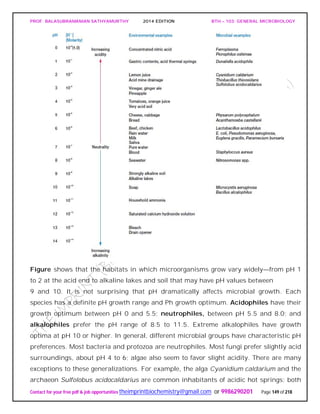
pH
pH is a measure of the hydrogen ion activity of a solution and is defined as the negative
logarithm of the hydrogen ion concentration (expressed in terms of molarity).
pH = - log [H+] = log(1/[H+])
The pH scale extends from pH 0.0 (1.0 M H+) to pH 14.0 (1.0 X 10-14 M H+), and each
pH unit represents a tenfold change in hydrogen ion concentration.](https://image.slidesharecdn.com/bth103generalmicrobiology-190421182551/85/Bth-103-general-microbiology-148-320.jpg)






![PROF. BALASUBRAMANIAN SATHYAMURTHY 2014 EDITION BTH – 103: GENERAL MICROBIOLOGY
Contact for your free pdf & job opportunities theimprintbiochemistry@gmail.com or 9986290201 Page 159 of 218
water, glassware, and regular media components often are adequate for growth.
Therefore it is very difficult to demonstrate a micronutrient requirement.
In nature, micronutrients are ubiquitous and probably do not usually limit growth.
Micronutrients are normally a part of enzymes and cofactors, and they aid in the
catalysis of reactions and maintenance of protein structure. For example, zinc (Zn2+) is
present at the active site of some enzymes but is also involved in the association of
regulatory and catalytic subunits in E. coli aspartate carbamoyltransferase.
Manganese (Mn2+) aids many enzymes catalyzing the transfer of phosphate groups.
Molybdenum (Mo2+) is required for nitrogen fixation, and cobalt (Co2+) is a component
of vitamin B12. Electron carriers and enzymes.
Besides the common macroelements and trace elements, microorganisms may have
particular requirements that reflect the special nature of their morphology or
environment.
Diatoms need silicic acid (H4SiO4) to construct their beautiful cell walls of silica
[(SiO2)n]. Although most bacteria do not require large amounts of sodium, many
bacteria growing in saline lakes and oceans depend on the presence of high
concentrations of sodium ion (Na+).
Finally, it must be emphasized that microorganisms require a balanced mixture of
nutrients. If an essential nutrient is in short supply, microbial growth will be limited
regardless of the concentrations of other nutrients.
Requirements for Carbon, Hydrogen, and Oxygen
The requirements for carbon, hydrogen, and oxygen often are satisfied together. Carbon
is needed for the skeleton or backbone of all organic molecules, and molecules serving
as carbon sources normally also contribute both oxygen and hydrogen atoms. They are
the source of all three elements. Because these organic nutrients are almost always
reduced and have electrons that they can donate to other molecules, they also can serve
as energy sources.
Indeed, the more reduced organic molecules are, the higher their energy content (e.g.,
lipids have higher energy content than carbohydrates). This is because, as we shall see
later, electron transfers release energy when the electrons move from reduced donors
with more negative reduction potentials to oxidized electron acceptors with more
positive potentials. Thus carbon sources frequently also serve as energy sources,
although they don’t have to.](https://image.slidesharecdn.com/bth103generalmicrobiology-190421182551/85/Bth-103-general-microbiology-159-320.jpg)









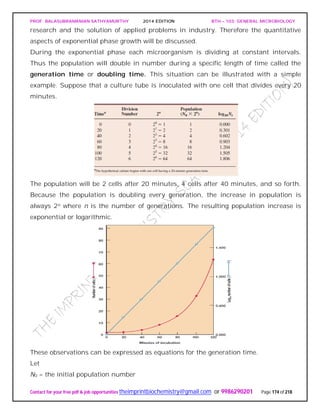

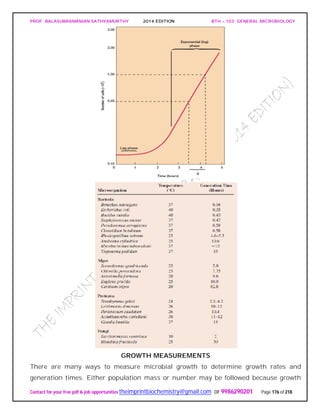









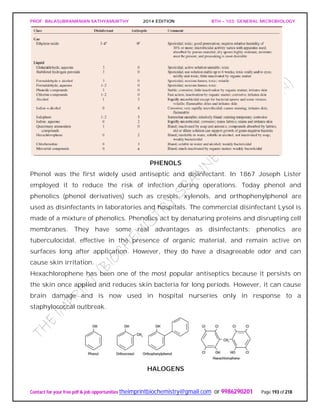

![PROF. BALASUBRAMANIAN SATHYAMURTHY 2014 EDITION BTH – 103: GENERAL MICROBIOLOGY
Contact for your free pdf & job opportunities theimprintbiochemistry@gmail.com or 9986290201 Page 195 of 218
chloride when added to water and disinfects it in about a half hour. It is frequently used
by campers lacking access to uncontaminated drinking water.
Chlorine solutions make very effective laboratory and household disinfectants. An
excellent disinfectant-detergent combination can be prepared if a 1/100 dilution of
household bleach (e.g., 1.3 fl oz of Clorox or Purex bleach in 1 gal or 10 ml/liter) is
combined with sufficient nonionic detergent (about 1 oz/gal or 7.8 ml/liter) to give a
0.8% detergent concentration. This mixture will remove both dirt and bacteria.
ALCOHOLS
Alcohols are among the most widely used disinfectants and antiseptics. They are
bactericidal and fungicidal but not sporicidal; some lipid-containing viruses are also
destroyed. The two most popular alcohol germicides are ethanol and isopropanol,
usually used in about 70 to 80% concentration. They act by denaturing proteins and
possibly by dissolving membrane lipids. A 10 to 15 minute soaking is sufficient to
disinfect thermometers and small instruments.
QUATERNARY AMMONIUM COMPOUNDS
Detergents [Latin detergere, to wipe off or away] are organic molecules that serve as
wetting agents and emulsifiers because they have both polar hydrophilic and nonpolar
hydrophobic ends. Due to their amphipathic nature, detergents solubilize otherwise
insoluble residues and are very effective cleansing agents. They are different than
soaps, which are derived from fats. Although anionic detergents have some
antimicrobial properties, only cationic detergents are effective disinfectants.](https://image.slidesharecdn.com/bth103generalmicrobiology-190421182551/85/Bth-103-general-microbiology-195-320.jpg)




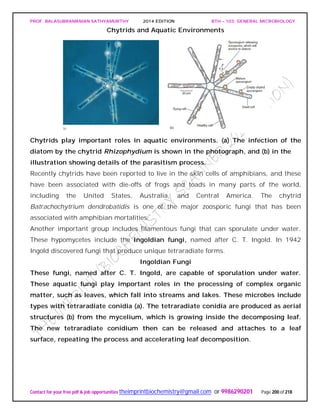
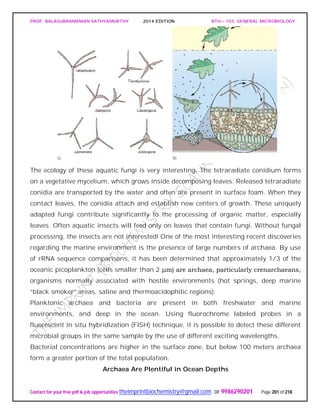
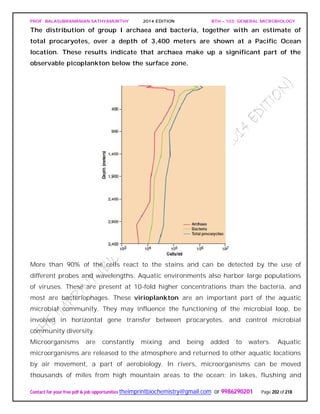

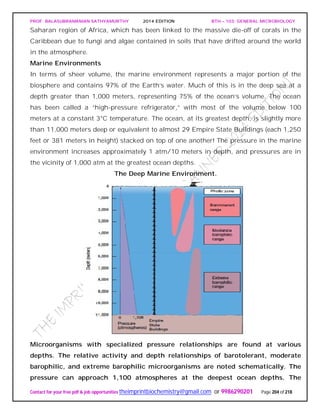
![PROF. BALASUBRAMANIAN SATHYAMURTHY 2014 EDITION BTH – 103: GENERAL MICROBIOLOGY
Contact for your free pdf & job opportunities theimprintbiochemistry@gmail.com or 9986290201 Page 205 of 218
depths are given in meters and “stacked” Empire State Buildings for perspective.
Light penetrates only into a relatively shallow surface layer, creating the photic
zone.
A series of pressure relationships is observed among the bacteria growing in this
vertically differentiated system. Some bacteria are barotolerant and grow between 0 and
400 atm, but best at normal atmospheric pressure. Many other bacteria are barophiles
[Greek, baro, weight and philein, to love] and prefer higher pressures. Moderate
barophiles grow optimally at 400 atm, but still grow at 1 atm; extreme barophiles grow
only at higher pressures. Pressure differences influence many biological processes
including cell division, flagellar assembly, DNA replication, membrane transport, and
protein synthesis. Porins, outer membrane proteins that form channels for diffusion of
materials into the periplasm, also function most effectively at specific pressures.
Most nutrient cycling in oceans occurs in the top 300 meters where light penetrates.
Light allows phytoplankton to grow and fall as a “marine snow” to the seabed. This
“trip” can take a month or longer. Most of the organic matter that falls below the 300
meter zone is decomposed, and only 1% of photosynthetically derived material reaches
the deep-sea floor unaltered. Because low inputs of organic matter occur in the deep
sea, the ability of microorganisms to grow under oligotrophic conditions becomes
important.
Freshwater Environments
Most fresh water that is not locked up in ice sheets, glaciers, or groundwaters is found
in lakes and rivers. These provide microbial environments that are different from the
larger oceanic systems in many important ways. For example, in lakes, mixing and
water exchange can be limited. This creates vertical gradients over much shorter
distances. Changes in rivers occur over distance and/or time as water flows through
river channels.
Lakes
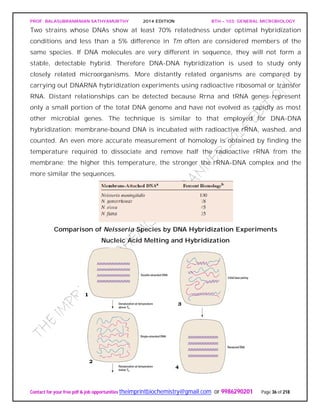
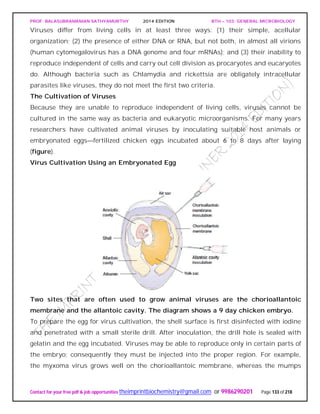
Lakes vary in nutrient status. Some are oligotrophic or nutrient poor, others are
eutrophic or nutrient-rich.(figureb).
Nutrient-poor lakes remain aerobic throughout the year, and seasonal temperature
shifts do not result in distinct oxygen stratification. In contrast, nutrient-rich lakes
usually have bottom sediments that contain organic matter. In thermally stratified lakes
the epilimnion (warm, upper layer) is aerobic, while the hypolimnion (deeper, colder,](https://image.slidesharecdn.com/bth103generalmicrobiology-190421182551/85/Bth-103-general-microbiology-205-320.jpg)