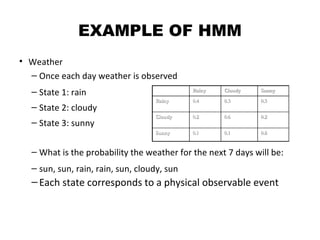
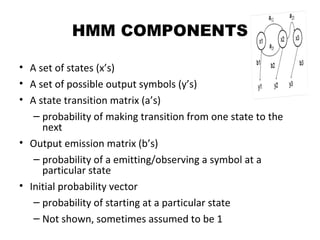
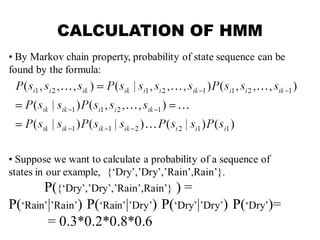
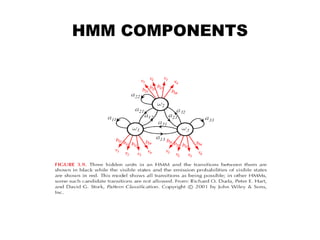
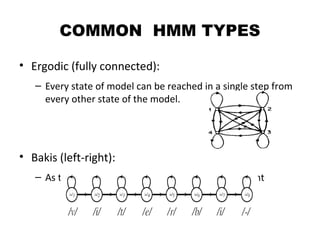
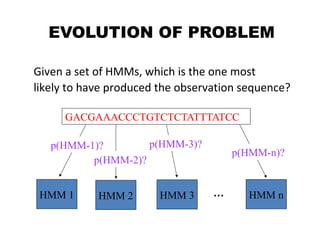
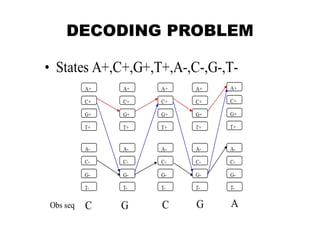
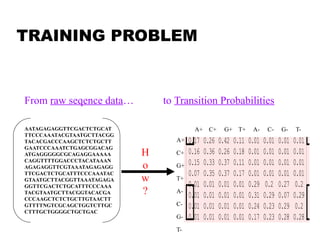
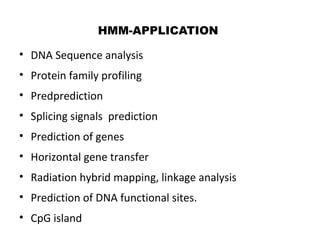
This document provides an introduction to hidden Markov models (HMMs). It discusses that HMMs can be used to model sequential processes where the underlying states cannot be directly observed, only the outputs of the states. The document outlines the basic components of an HMM including states, transition probabilities, emission probabilities, and gives examples of HMMs for coin tossing and weather. It also briefly discusses the history of HMMs and their applications in fields like bioinformatics for problems such as gene finding.