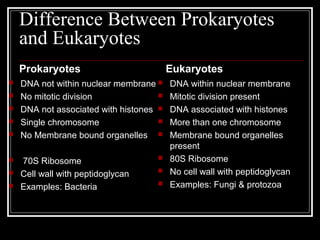
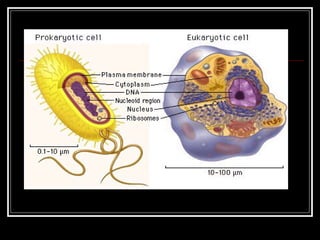
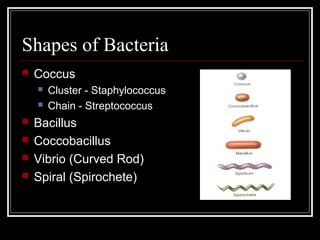
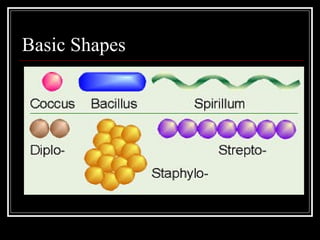
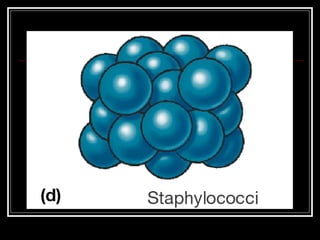
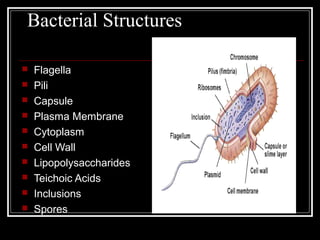
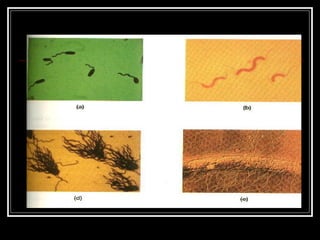
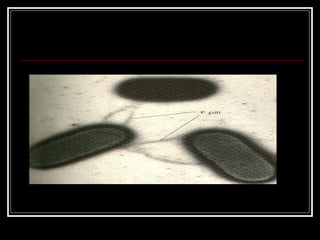
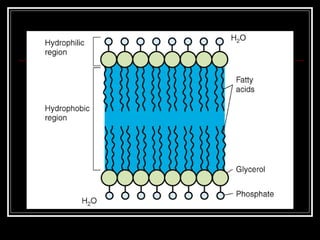
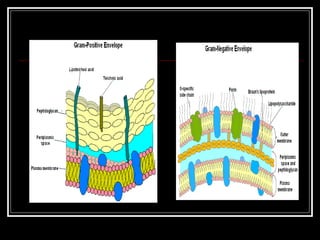
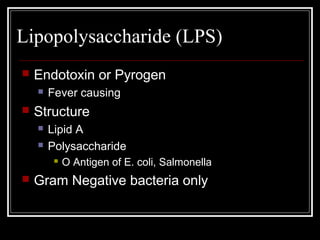
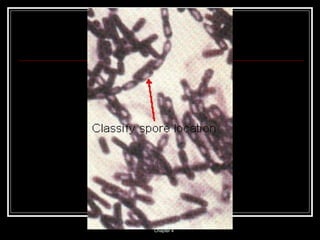
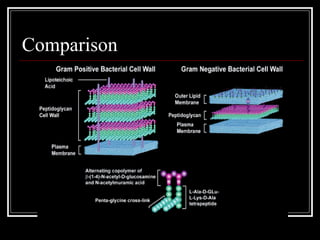
This document discusses the structure of bacteria. It begins by comparing prokaryotes and eukaryotes, noting key differences such as DNA location, organelles, and ribosomes. It then describes bacteria specifically, including their size, shapes, structures like flagella and cell walls, and differences between gram positive and gram negative bacteria. Important bacterial components are also outlined, such as peptidoglycan, teichoic acids, lipopolysaccharides, and endospores.