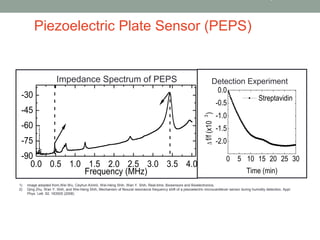
1) A method was developed to detect transrenal DNA mutations using a piezoelectric plate sensor with attomolar sensitivity and specificity to detect mutant DNA in a background of wild type DNA.
2) Signal processing algorithms and a moving window approach were optimized to reduce noise and accurately detect mutation peaks with concentrations as low as 60 copies/ml.
3) Laminar flow and temperature conditions were identified that maximized specificity for detecting mixtures of mutant and wild type target DNA.
4) The method was validated through two-color fluorescent microsphere reporter hybridization and applied to detect double-stranded target DNA and DNA extracted from cell cultures.









![Sensitivity : Noise problem in data
processing
• Inability to detect low concentrations of analytes due to
noise in data.
Reason
1.Imprecise peak
finding algorithm.
2.Stationary window
losing important data
while peak shifts.
0 5 10 15 20 25 30
-2
-1
0
1
2
FrequencyChange(kHz)
Time (min)
[tDNA]=10
-19
M
Control
11](https://image.slidesharecdn.com/presentation-151228001420/85/Piezoelectric-Plate-Sensor-11-320.jpg)




































