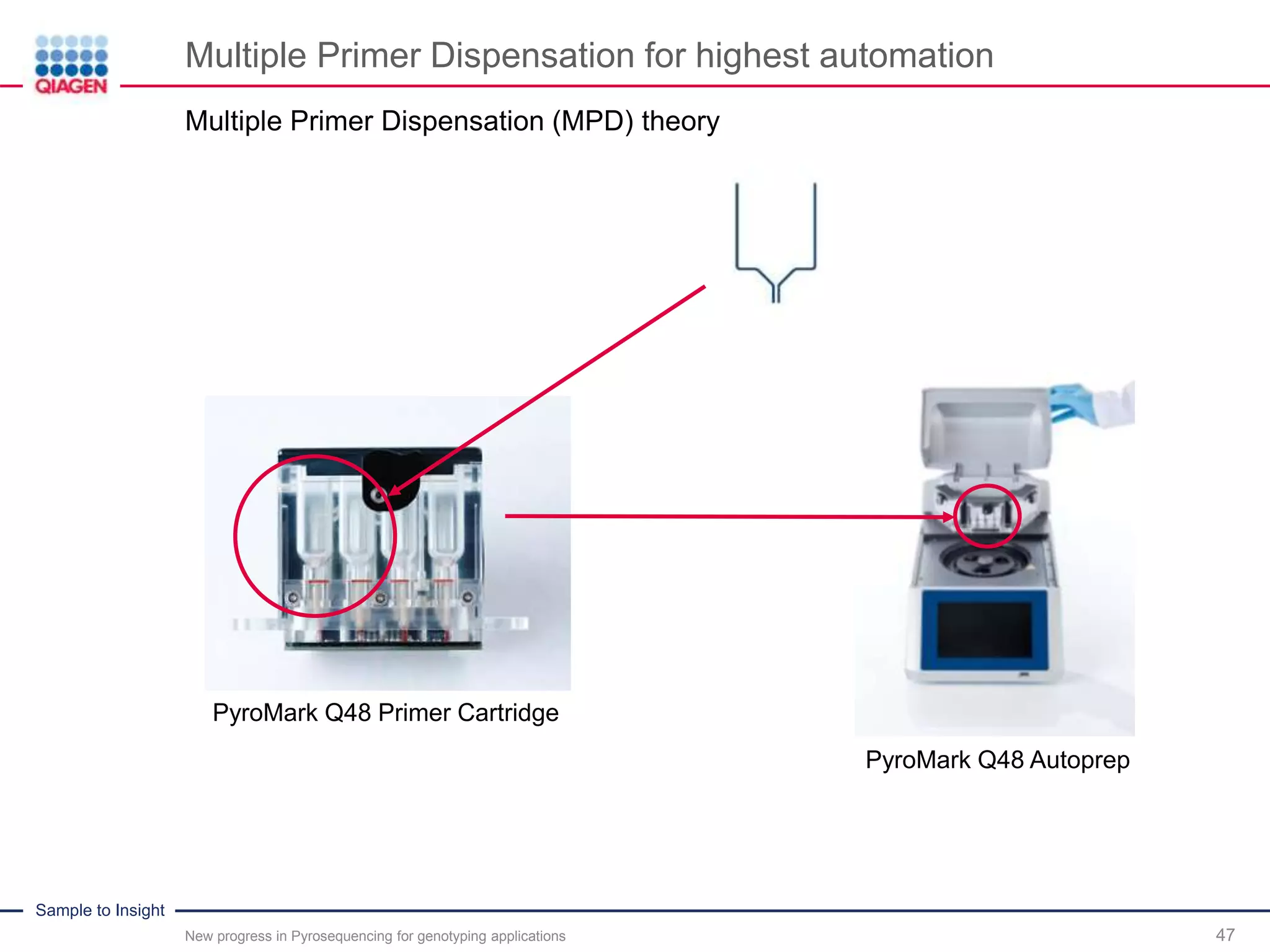
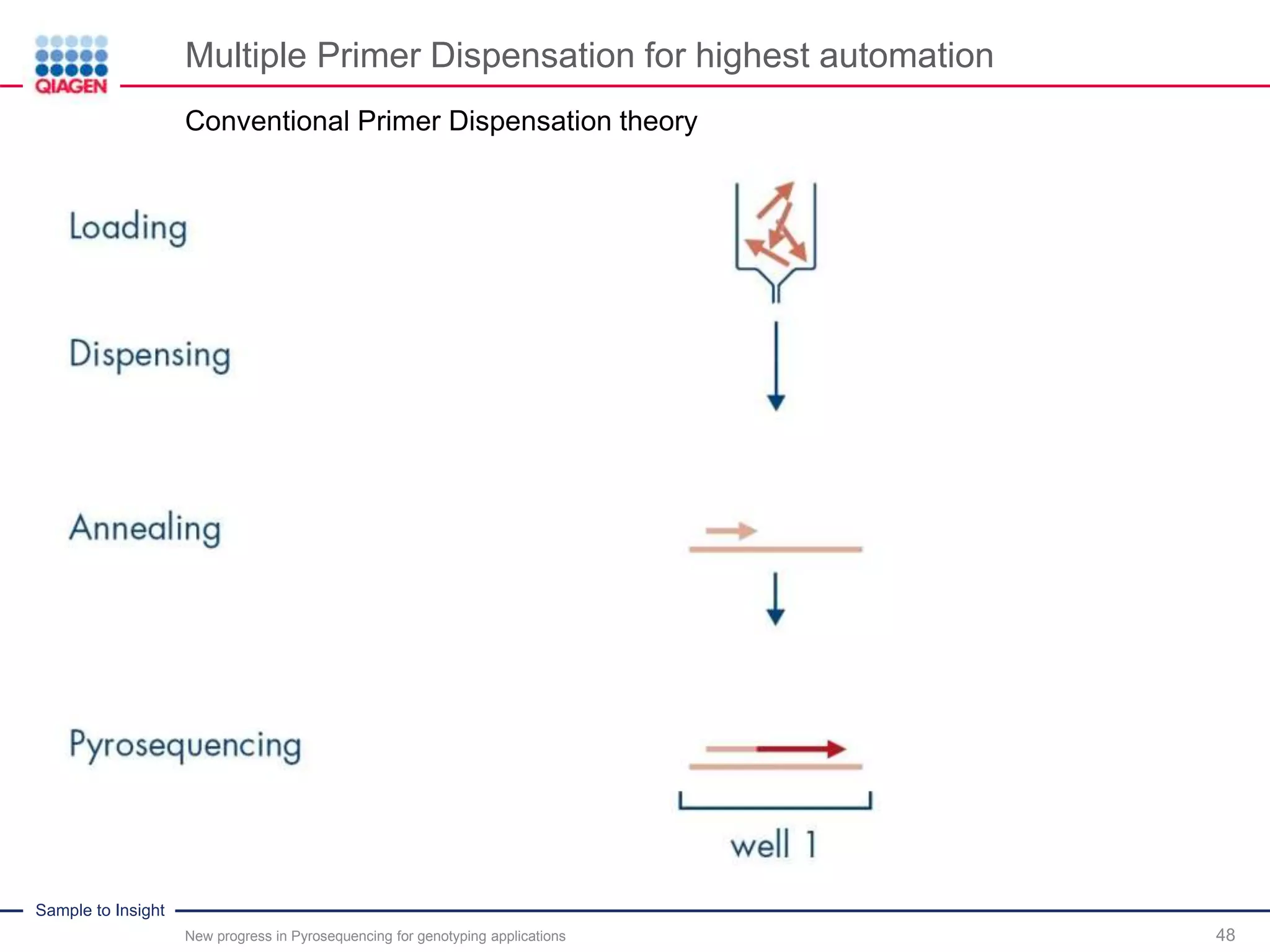
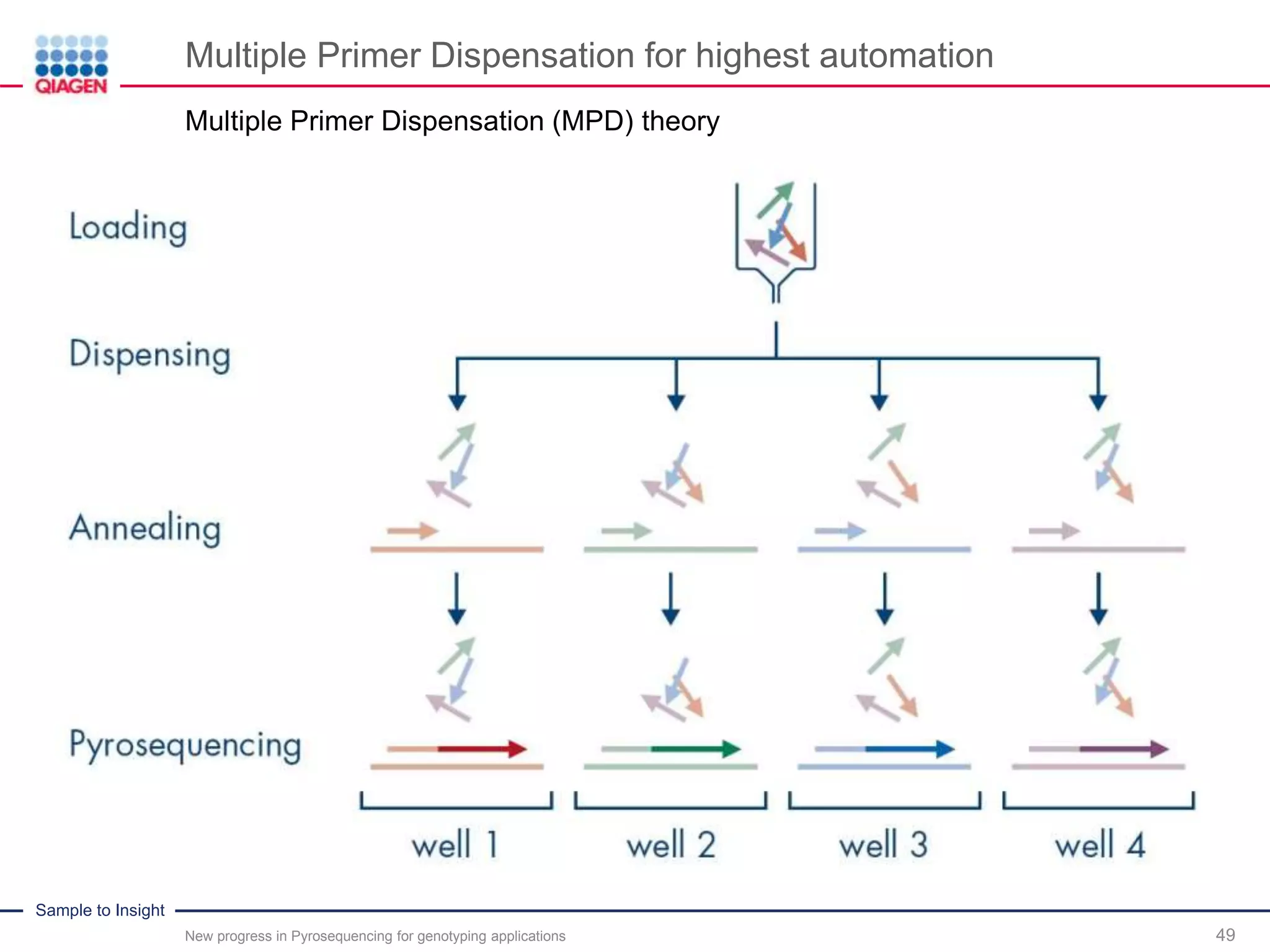
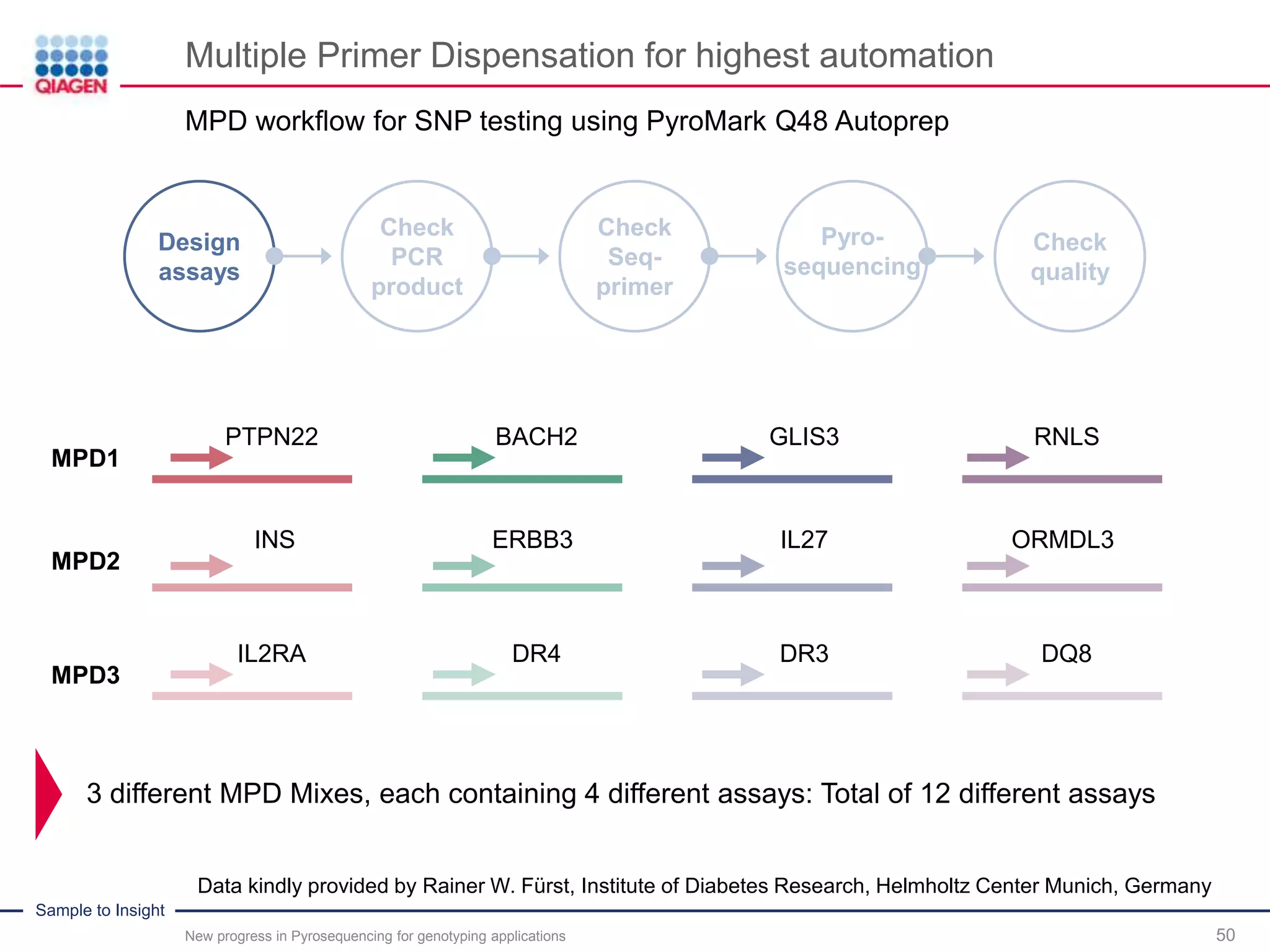
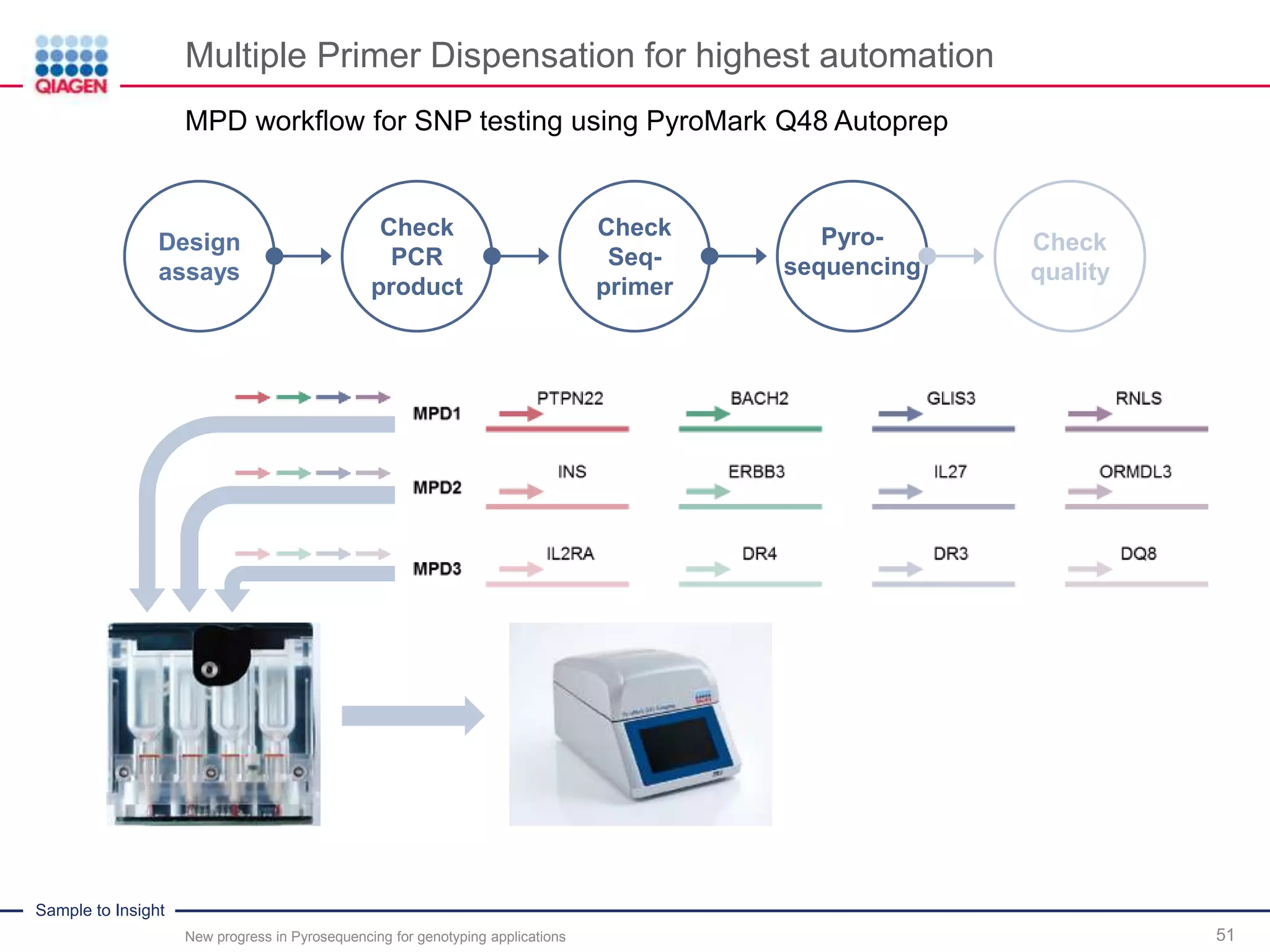
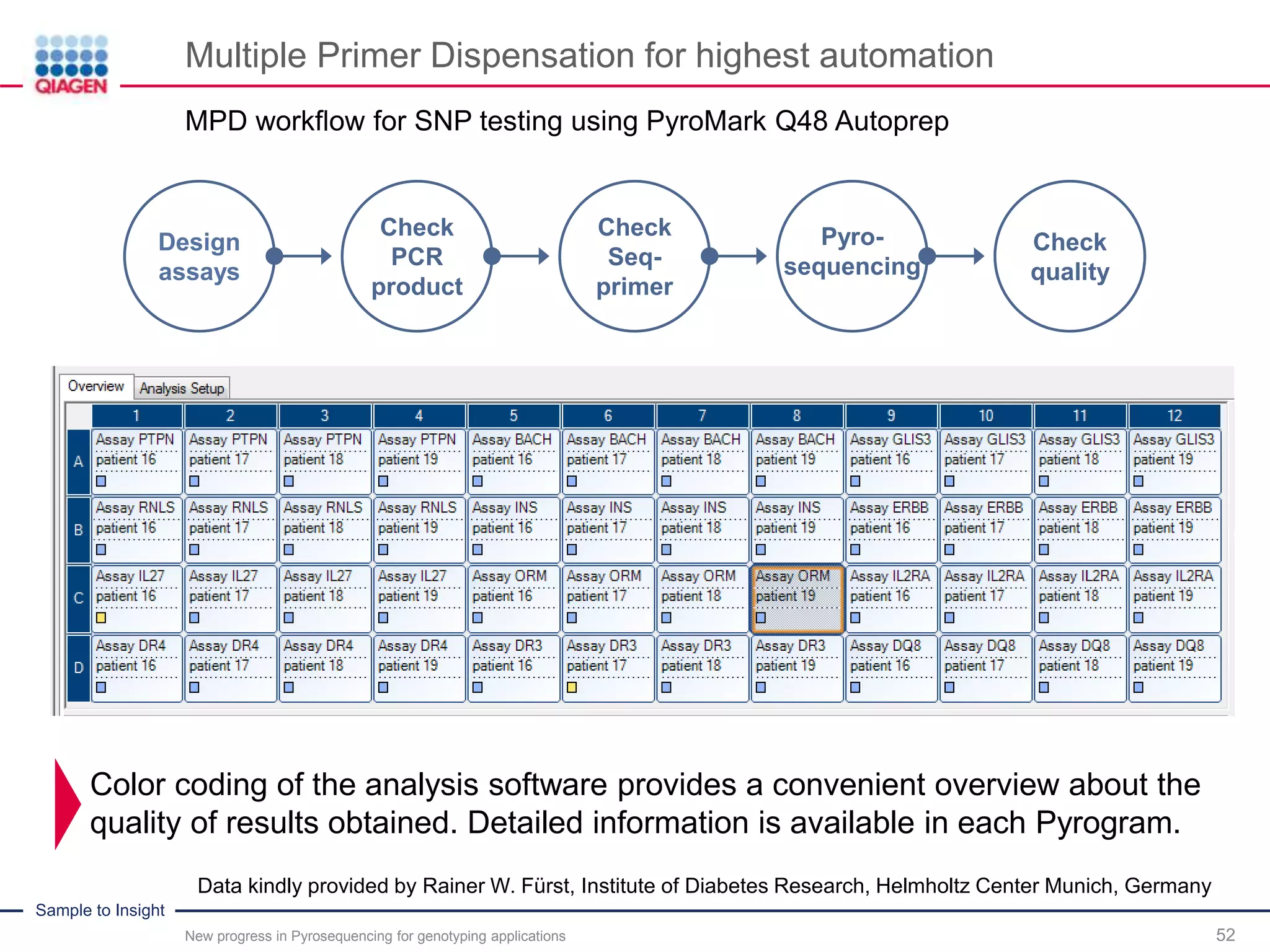
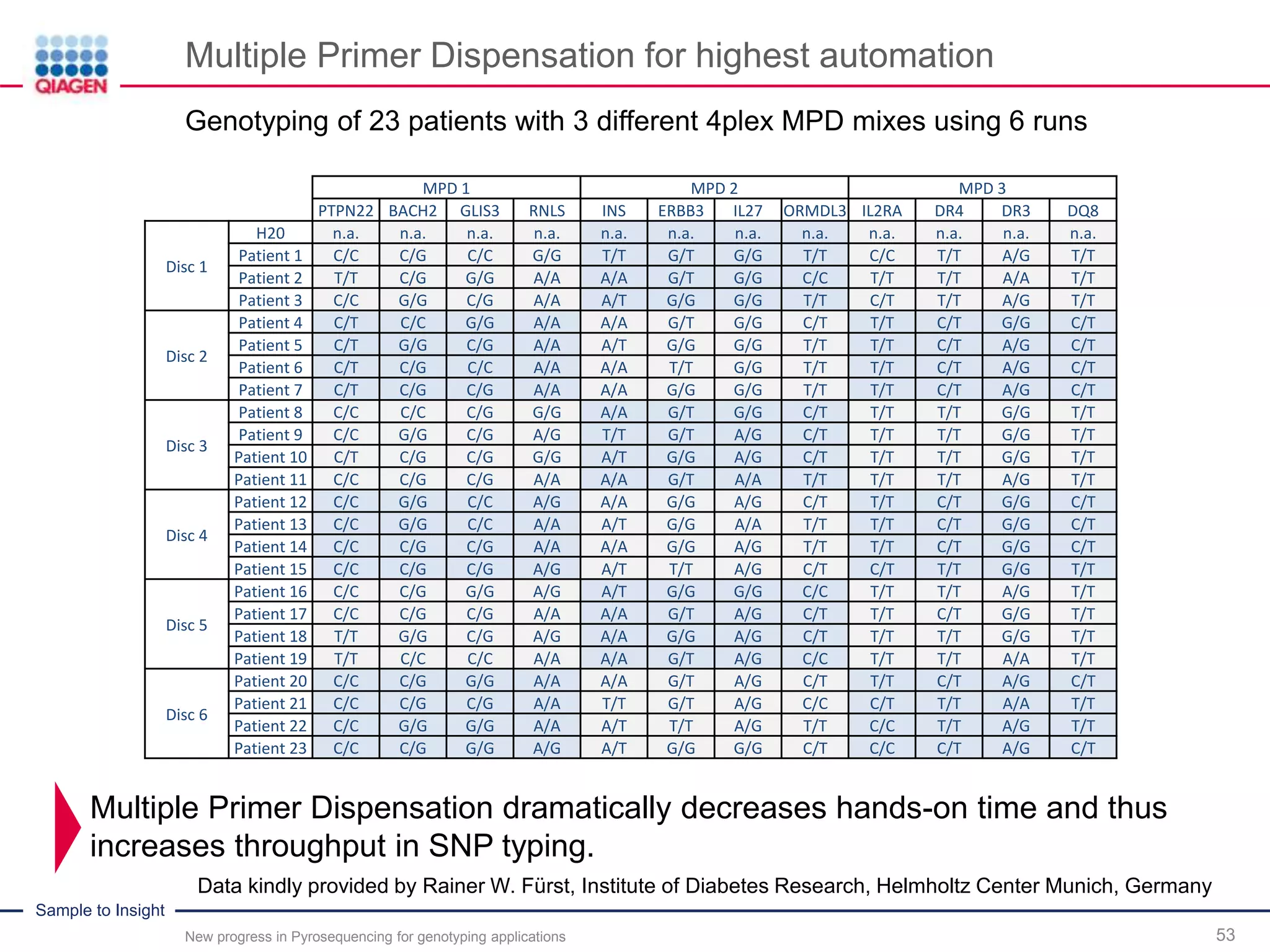
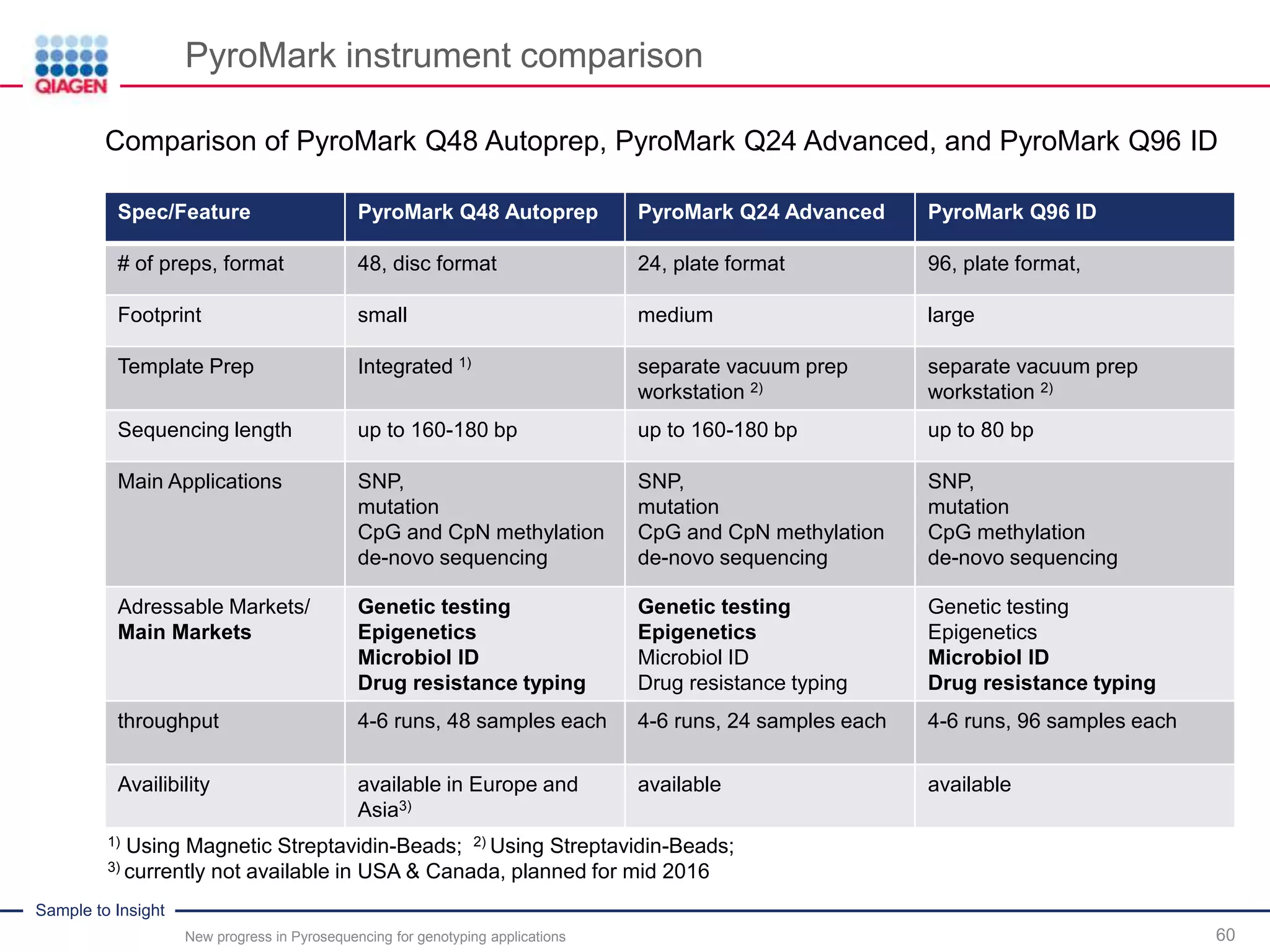
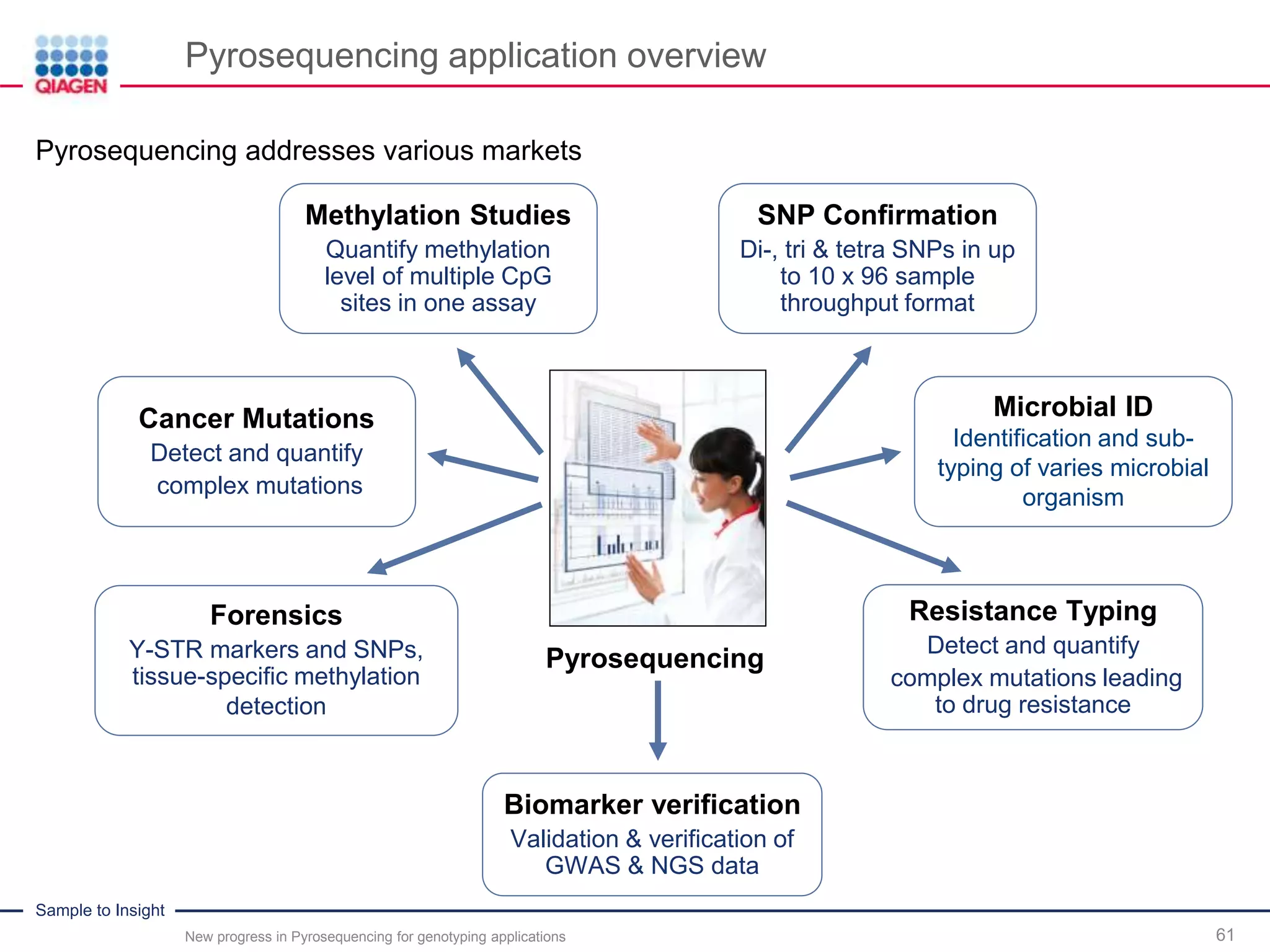
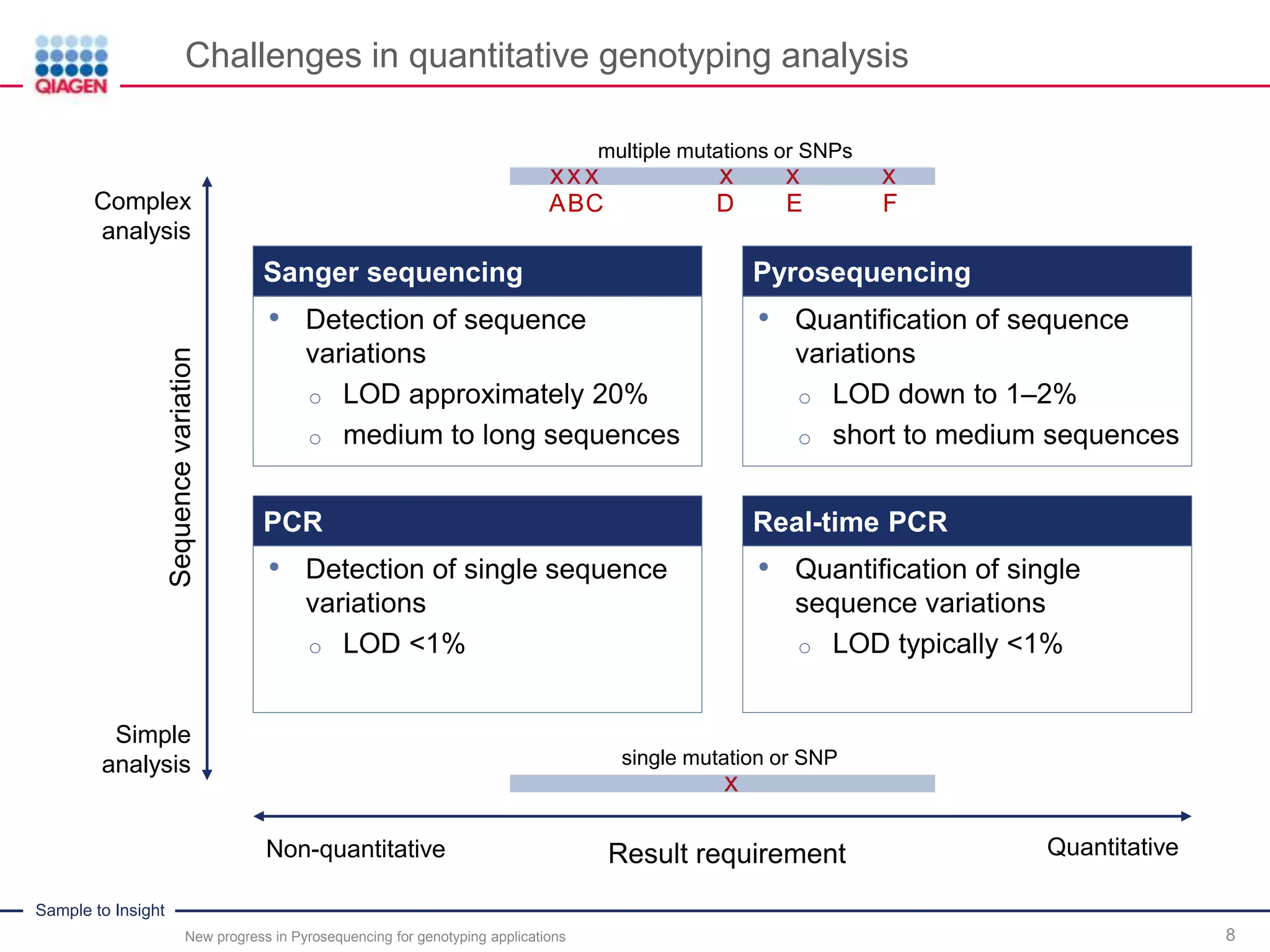
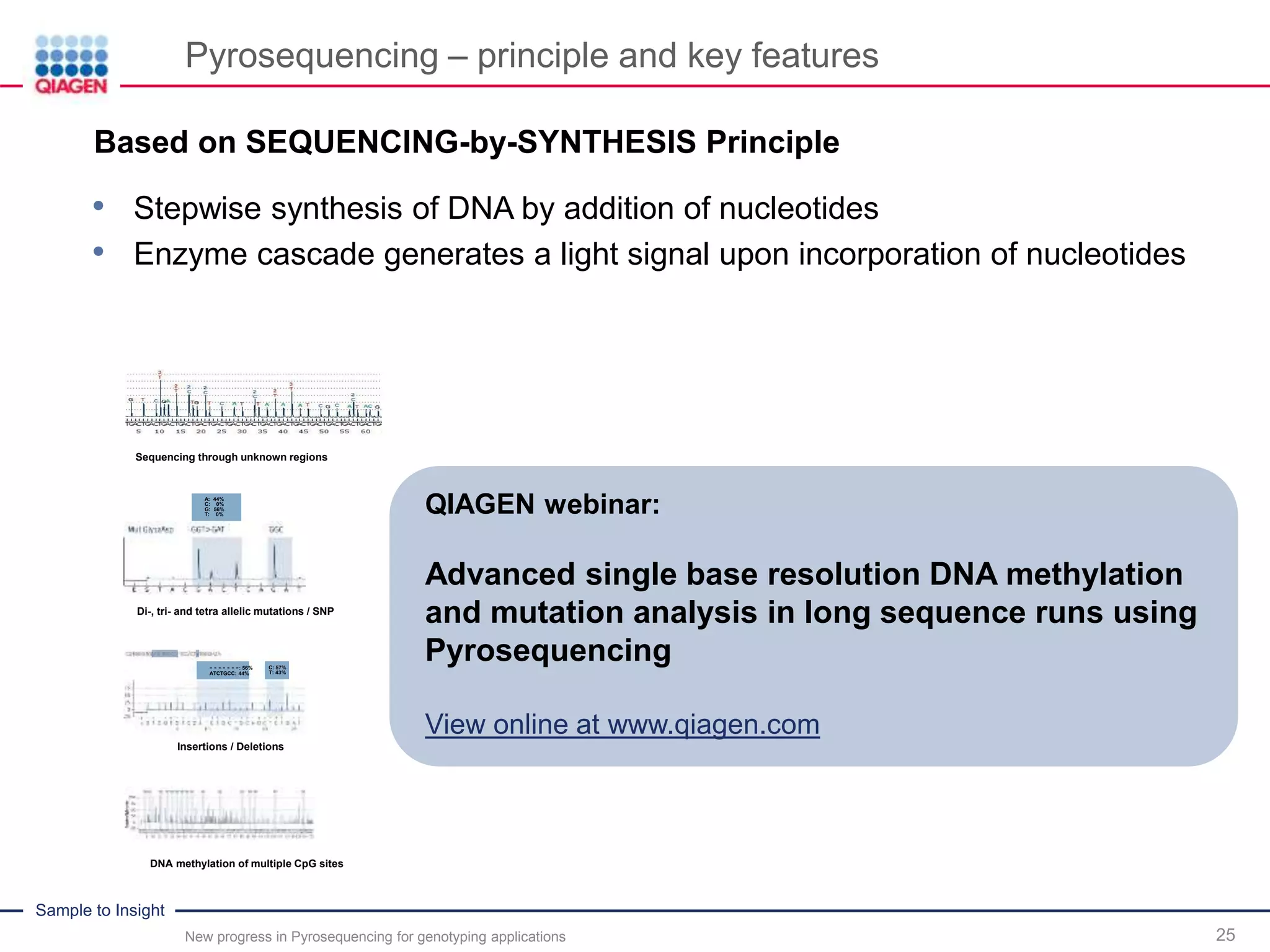
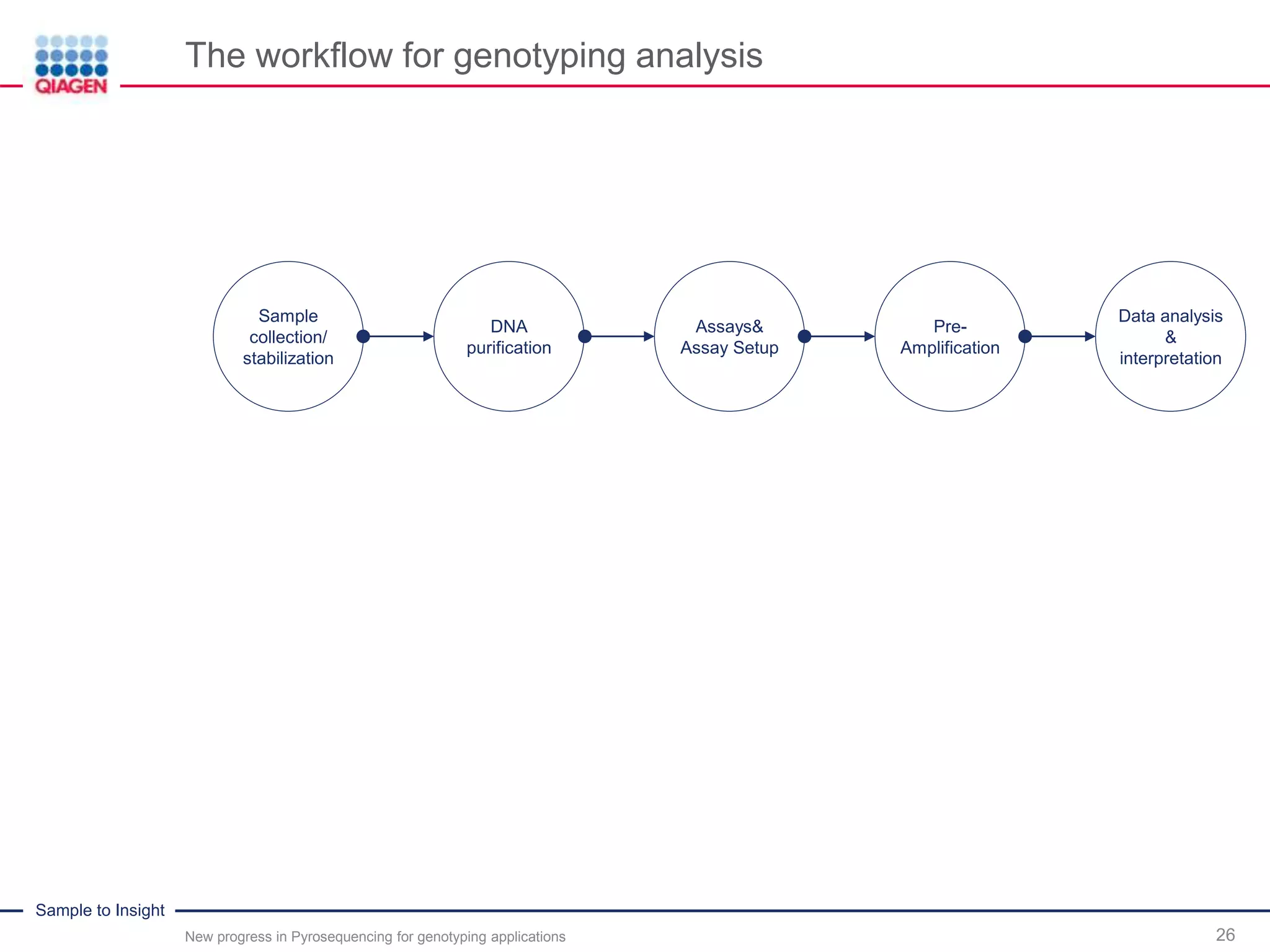
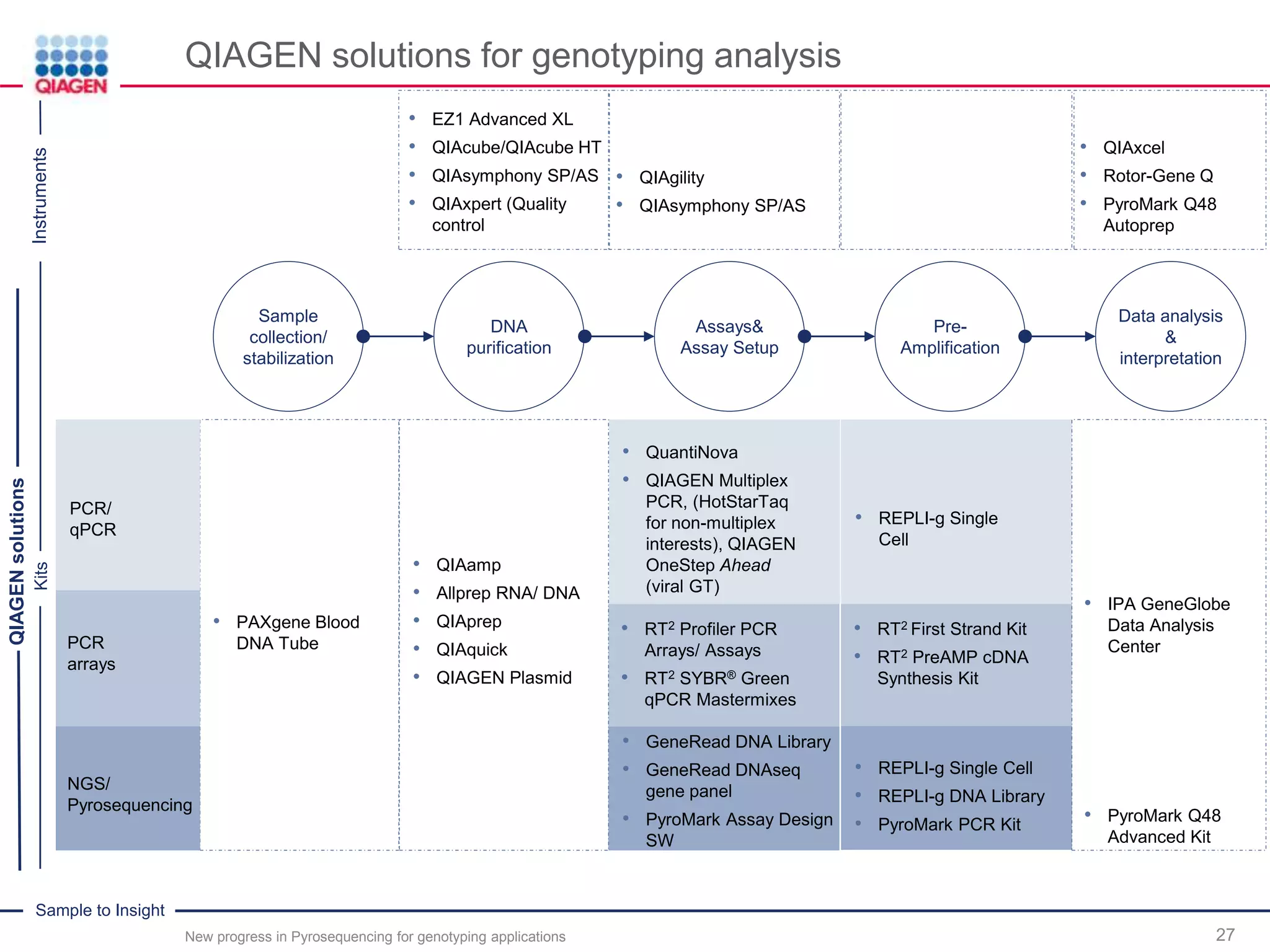
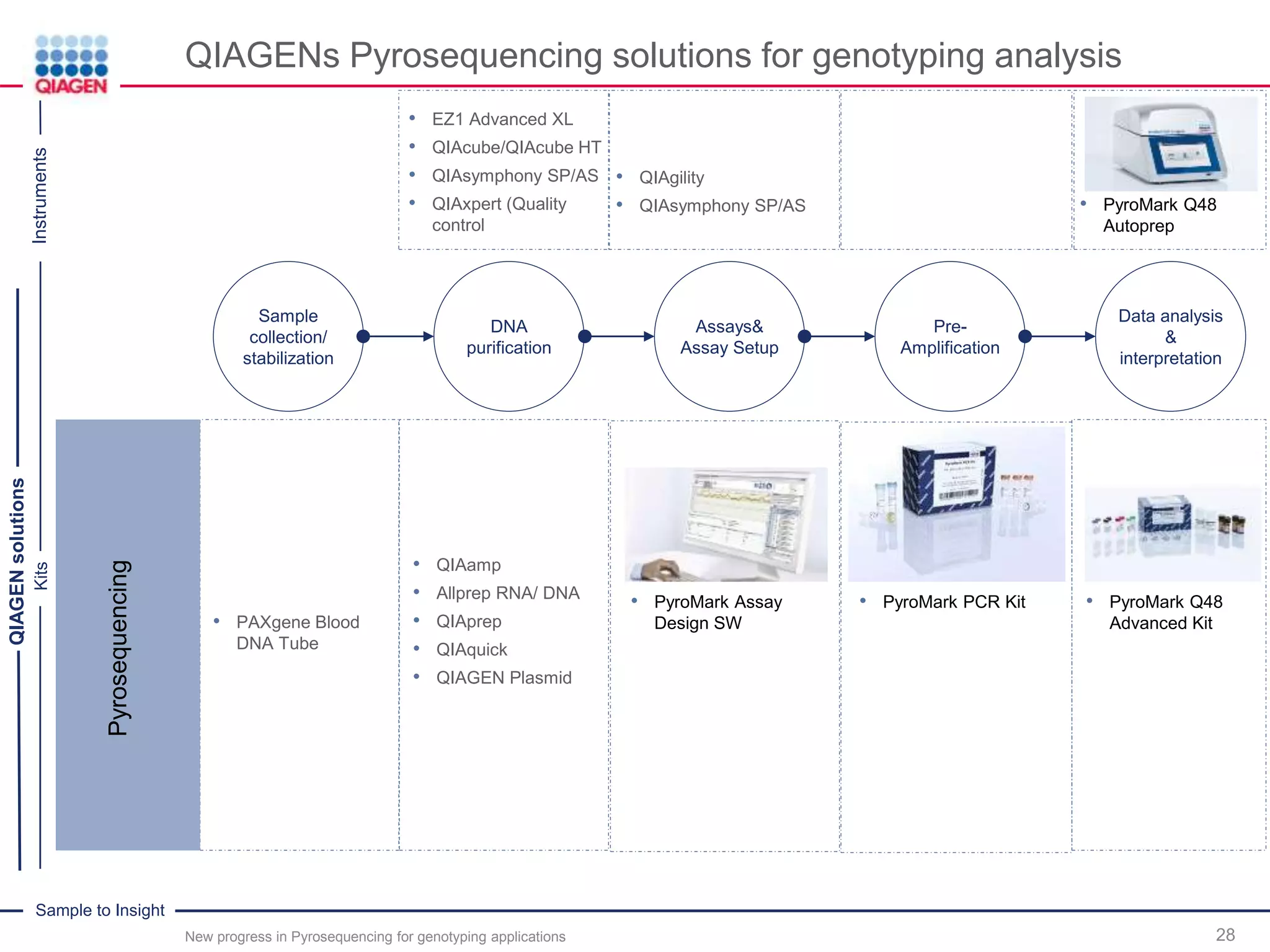
The document discusses advancements in pyrosequencing technology for automated quantitative analysis of bi- or multi-allelic sequence variations, emphasizing its application in genotyping. It outlines challenges in quantitative genotyping analysis and presents the Pyromark Q48 Autoprep MPD strategy to enhance workflow efficiency. Additionally, it provides an overview of product offerings, assay design, and the sequencing process itself.

![Sample to Insight
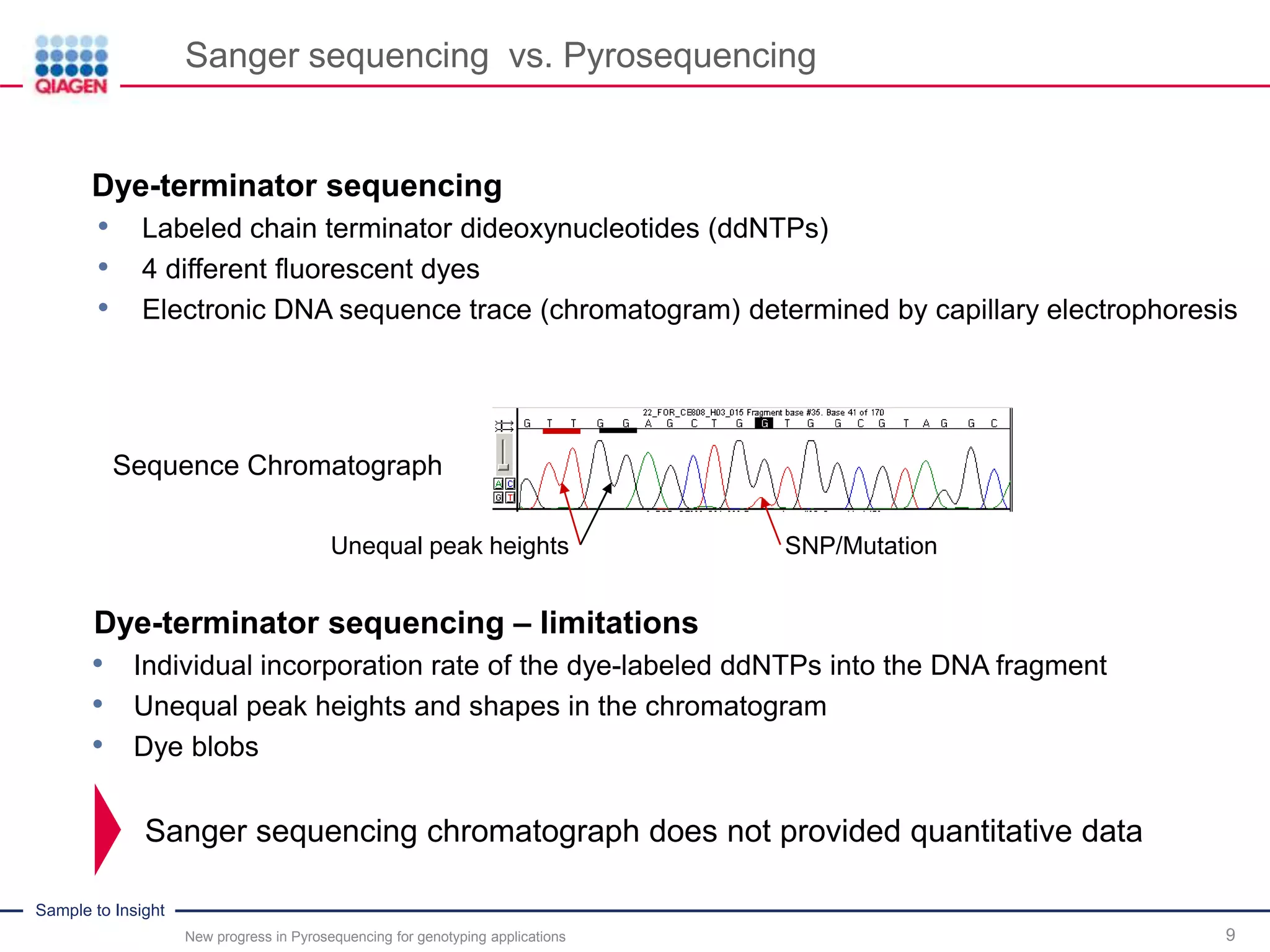
Sequence Chromatograph
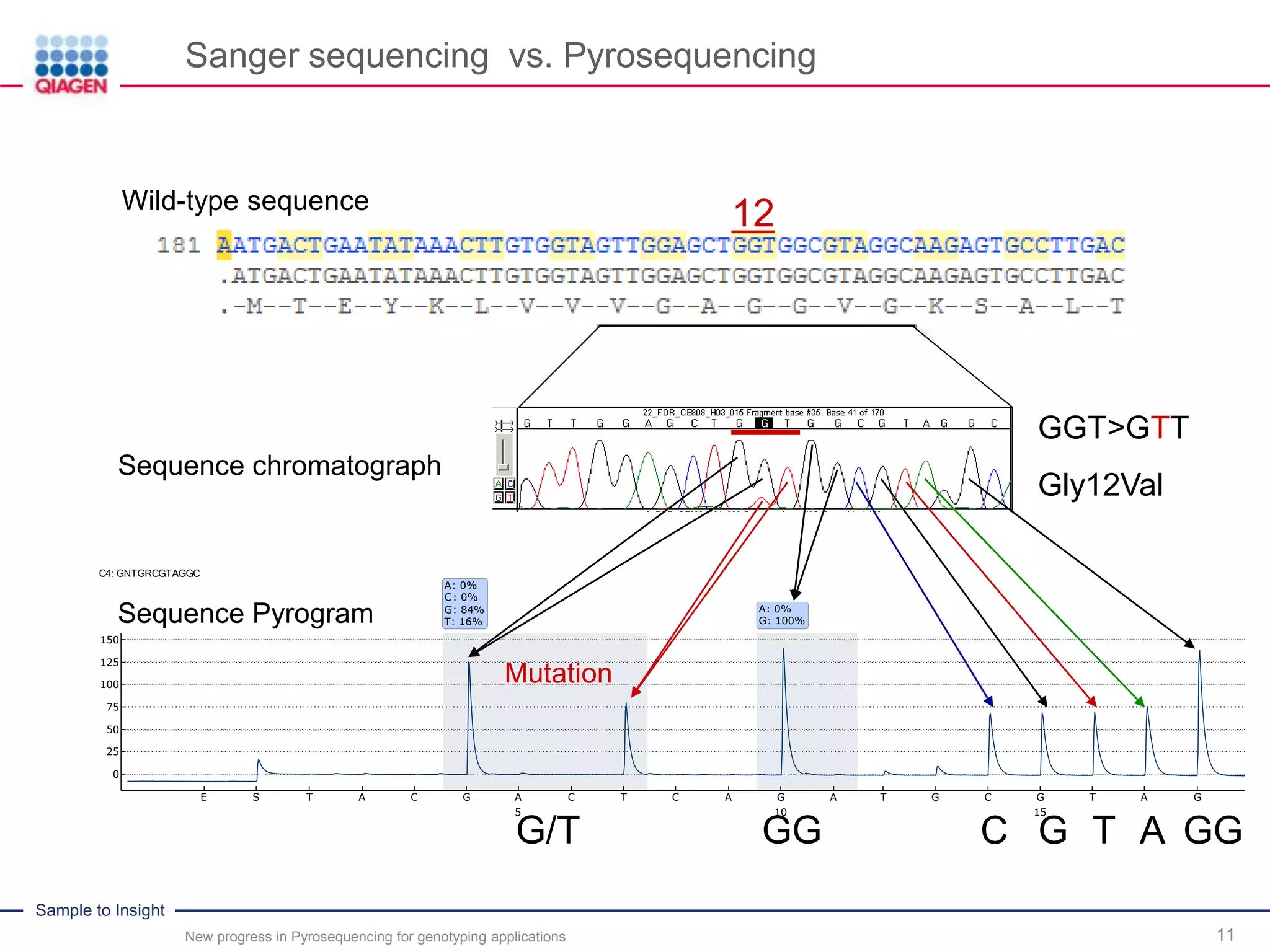
Sanger sequencing vs. Pyrosequencing
New progress in Pyrosequencing for genotyping applications 10
Dye-terminator sequencing
• Labeled chain terminator dideoxynucleotides (ddNTPs)
• 4 different fluorescent dyes
• Electronic DNA sequence trace (chromatogram) determined by capillary electrophoresis
Unequal peak heights SNP/Mutation
Nickel, G.C. et al. Characterizing Mutational Heterogeneity in a Glioblastoma Patient with Double Recurrence. PLoS
One, 2012
“… The signals from such mutations [low frequency mutations] are often below the
noise threshold in Sanger sequence reads, … it is crucial that researchers do not rely
solely on capillary-based sequencing for mutation detection and validation...”](https://image.slidesharecdn.com/prom-9583-001newprogressinpyrosequencingwebinar0416ww-160502142235/75/New-Progress-in-Pyrosequencing-for-Automated-Quantitative-Analysis-of-Bi-or-Multi-allelic-Sequence-Variations-Webinar-10-2048.jpg)
![Sample to Insight
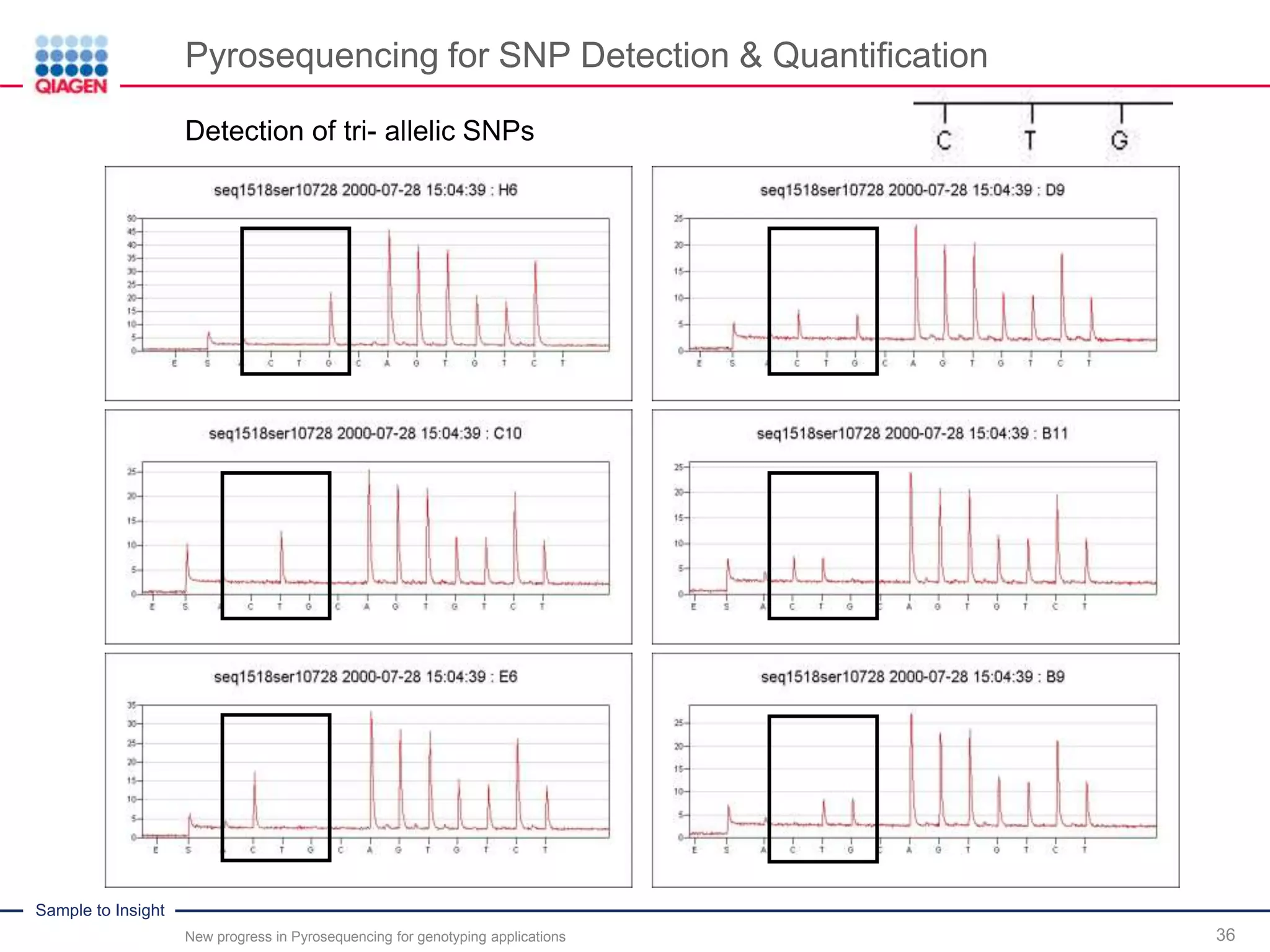
Pyrosequencing for SNP Detection & Quantification
New progress in Pyrosequencing for genotyping applications 38
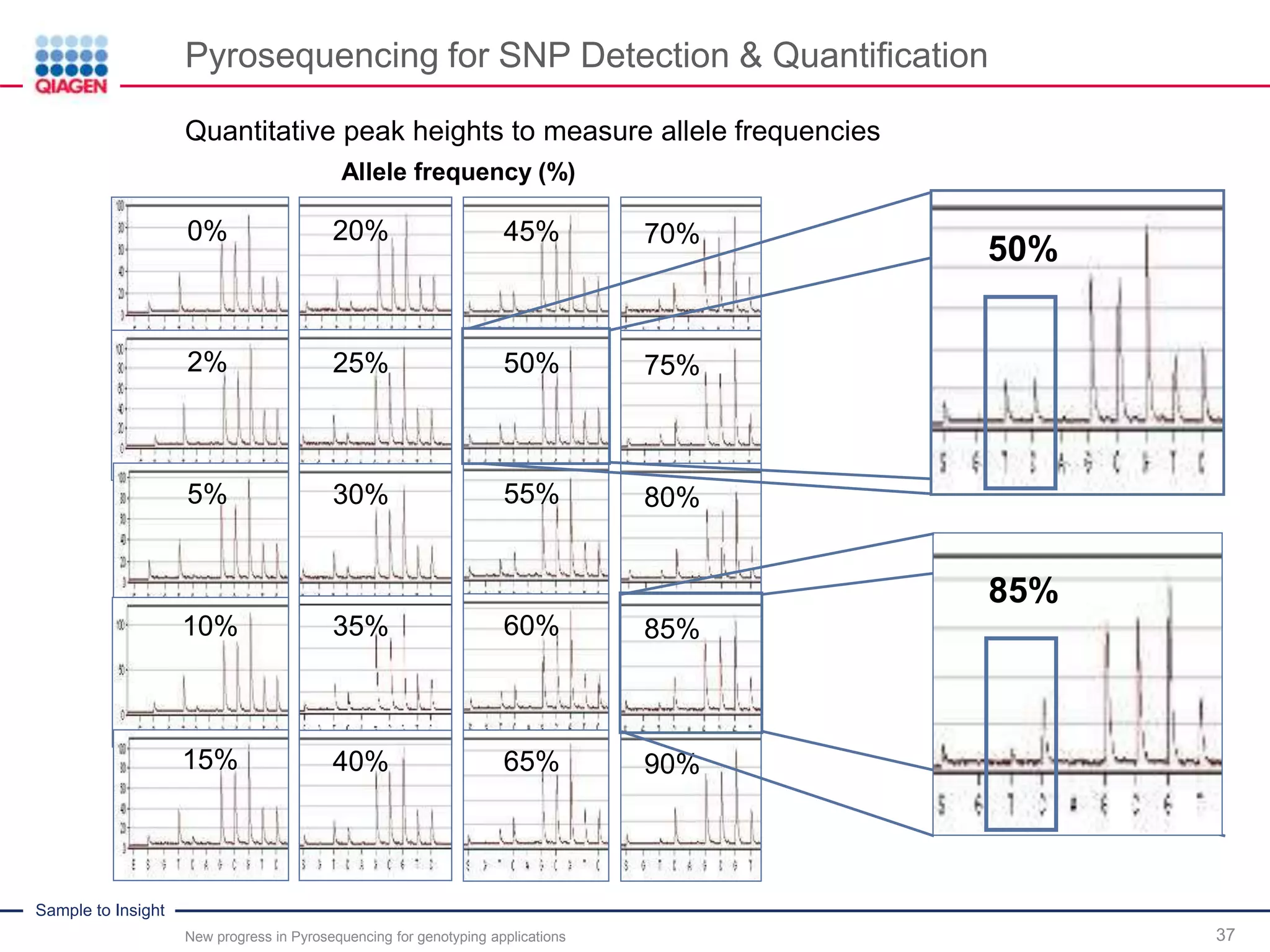
Quantitative peak heights to measure allele frequencies
Quantitative peak heights
R2
= 0.9993
0.0
0.1
0.2
0.3
0.4
0.5
0.6
0.7
0.8
0.9
1.0
0 5 10 15 20 25 30 35 40 45 50 55 60 65 70 75 80 85 90 95 100
Allele frequency [%]
Rel.peakheight
SNP E1
Linear (SNP E1)
Even as little as 2% of one allele
in 98% of the other could be detected
50%
85%](https://image.slidesharecdn.com/prom-9583-001newprogressinpyrosequencingwebinar0416ww-160502142235/75/New-Progress-in-Pyrosequencing-for-Automated-Quantitative-Analysis-of-Bi-or-Multi-allelic-Sequence-Variations-Webinar-38-2048.jpg)