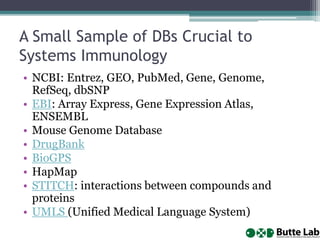
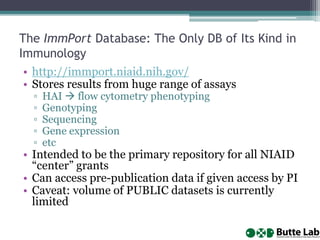
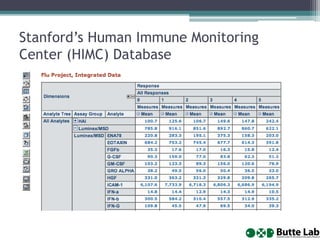
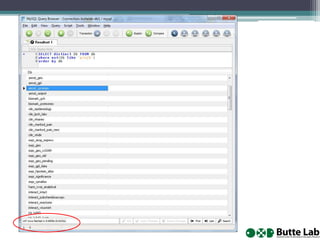
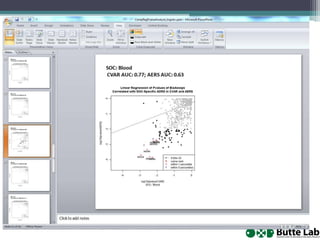
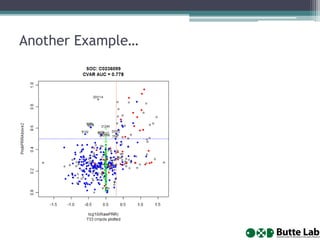
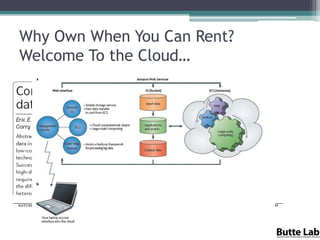
This document provides an overview of databases, web services, tools, and computing resources needed for systems immunology. It discusses the importance of having a clear hypothesis, statistical understanding, large datasets from different levels of biology, software tools, programming expertise, and computing power. Specific databases, tools, and programming languages discussed include ImmPort, Stanford's HIMC database, MySQL, GenePattern, Galaxy, Weka, R, Perl, Python, and Amazon Cloud computing. The document provides recommendations and resources for learning statistics, data mining, programming languages, and using cloud computing resources.