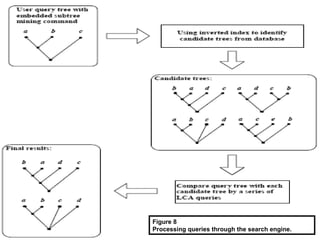
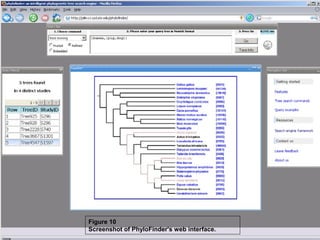
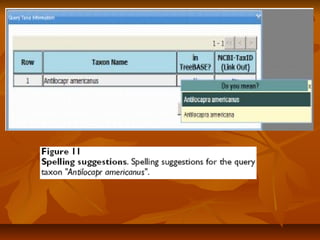
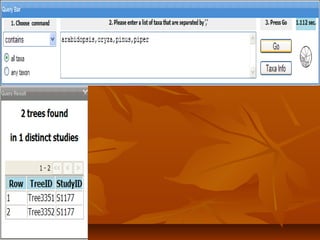
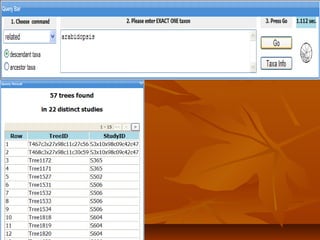
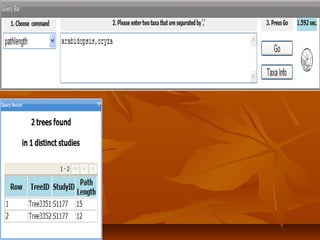
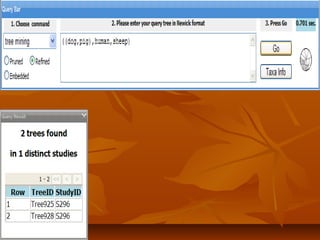
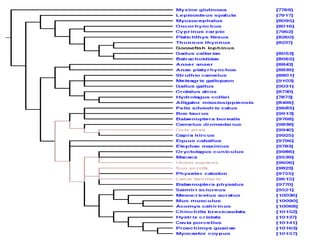
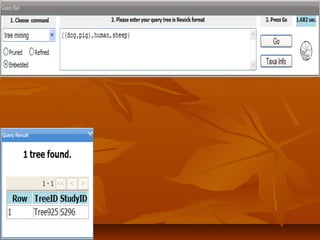
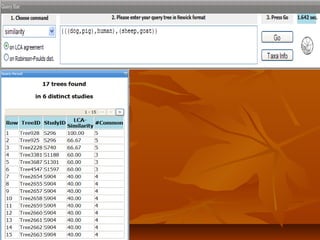
Phylofinder is a search engine designed to enhance access to phylogenetic tree databases, specifically addressing challenges such as taxonomic inconsistencies and naming changes. It utilizes tbmap to interpret synonymous taxon names and includes a visualization tool for query results. The system supports both taxonomic and phylogenetic queries to locate and compare trees within a comprehensive database of over 4,400 phylogenetic trees.



















![System ArchitectureSystem Architecture
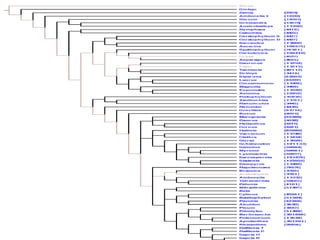
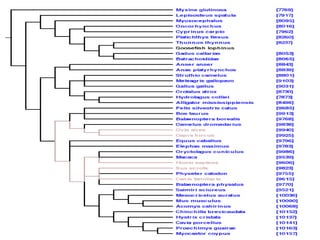
PhyloFinder stores the phylogenetic trees, which in our test implementationPhyloFinder stores the phylogenetic trees, which in our test implementation
are from TreeBASE, and the NCBI taxonomy tree in MySQL (an openare from TreeBASE, and the NCBI taxonomy tree in MySQL (an open
source relational database management system (RDBMS)) using a slightsource relational database management system (RDBMS)) using a slight
modification ofmodification of nested-set representationnested-set representation..
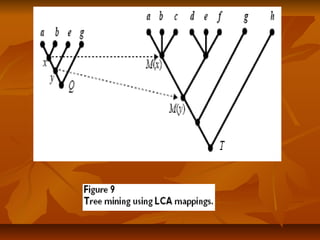
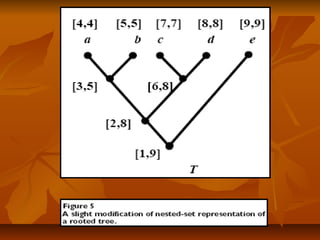
Under this scheme, each nodeUnder this scheme, each node xx of a given tree is represented by an intervalof a given tree is represented by an interval
[[NxNx,, RxRx], where], where Nx (Nx ( NodeID ofNodeID of x)x), is an integer defined by a preorder walk, is an integer defined by a preorder walk
of the tree, andof the tree, and RxRx is the largest NodeID of a descendant ofis the largest NodeID of a descendant of xx (Figure 5).(Figure 5).
NodeNode yy is an ancestor of nodeis an ancestor of node xx if and only if the interval [if and only if the interval [NyNy,, RyRy] contains] contains
the interval [the interval [Nx, RxNx, Rx], and the LCA of a set of nodes], and the LCA of a set of nodes xx1,...,1,...,xkxk is the commonis the common
ancestorancestoryy with largest NodeID. With this representation, LCA queries canwith largest NodeID. With this representation, LCA queries can
be implemented as SQL queries directly on the relational database.be implemented as SQL queries directly on the relational database.
Taxon clusters are generated using TBMap and the NCBI taxonomyTaxon clusters are generated using TBMap and the NCBI taxonomy
database.database.](https://image.slidesharecdn.com/phylofinder-171015215034/85/Phylo-finder-an-intelligent-search-engine-for-phylogenetic-tree-databases-33-320.jpg)



![System ArchitectureSystem Architecture
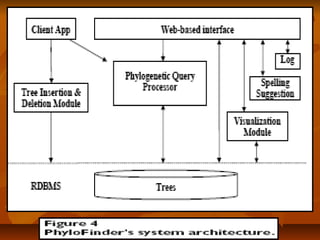
TheThe client applicationclient application is an administration tool that provides an interface tois an administration tool that provides an interface to
handle tree insertions and deletions (via thehandle tree insertions and deletions (via the insertion and deletion moduleinsertion and deletion module).).
It also provides commands for updating the inverted index and performing low-It also provides commands for updating the inverted index and performing low-
level queries (SQL commands or specified query commands with unformattedlevel queries (SQL commands or specified query commands with unformatted
results) for testing and debugging.results) for testing and debugging.
TheThe web-based interfaceweb-based interface is an interactive web application that uses the AJAXis an interactive web application that uses the AJAX
technique. It is written using the Google Web Toolkit (GWT) [27] and the GWTtechnique. It is written using the Google Web Toolkit (GWT) [27] and the GWT
Window Manager (GWM).Window Manager (GWM).
The system maintains aThe system maintains a loglog, where it records user queries and timestamps., where it records user queries and timestamps.
The statistics can be used to analyze user needs and help optimize the searchThe statistics can be used to analyze user needs and help optimize the search
engine performance.engine performance.](https://image.slidesharecdn.com/phylofinder-171015215034/85/Phylo-finder-an-intelligent-search-engine-for-phylogenetic-tree-databases-39-320.jpg)

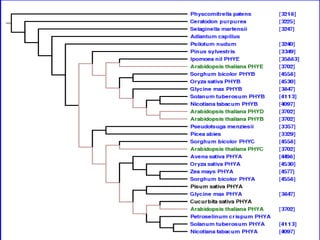
![Figure 7
Synonym conflict in IPNI and
NCBI. Name cluster containing
"Antennaria solitaria" and
"Antennaria monocephala"
produced
by TBMap [8]. Anternnaria
solitaria and Antennaria
monocephala are synonyms in
IPNI, but are treated as distinct
taxa by NCBI.
In such cases, PhyloFinder uses
the NCBI classification.](https://image.slidesharecdn.com/phylofinder-171015215034/85/Phylo-finder-an-intelligent-search-engine-for-phylogenetic-tree-databases-41-320.jpg)