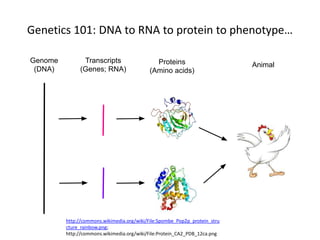
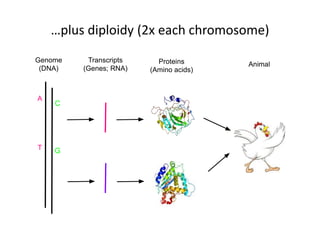
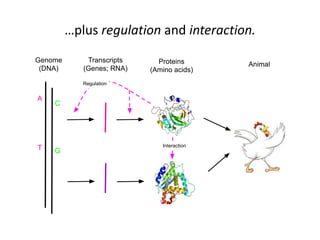
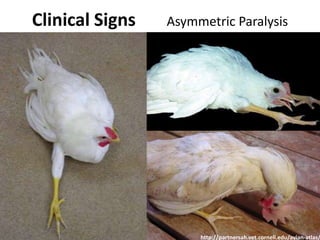
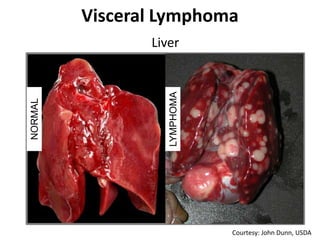
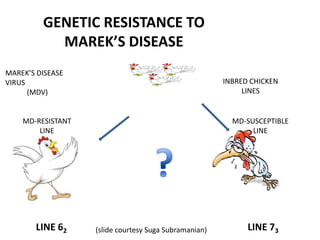
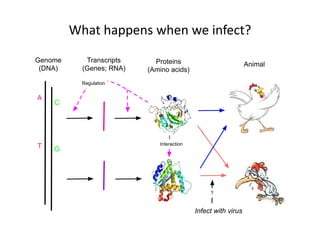
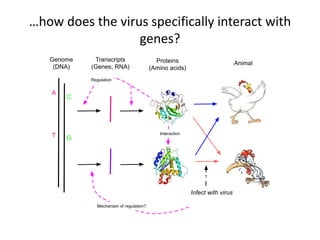
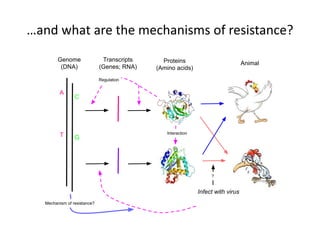
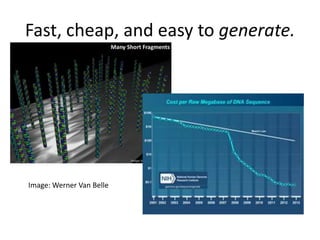
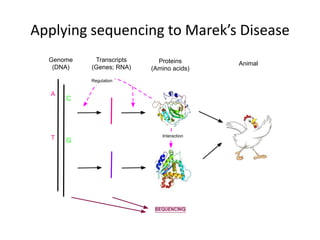
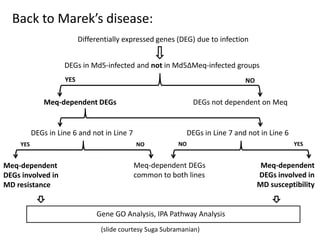
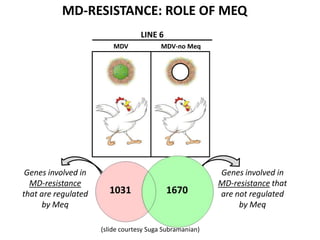
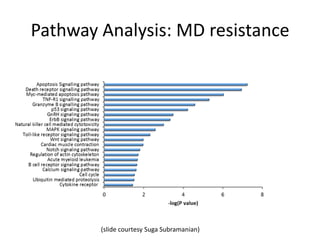
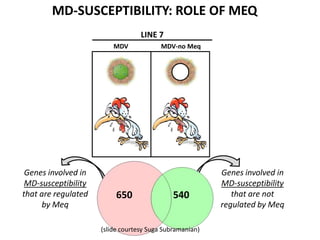
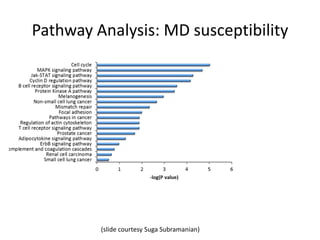
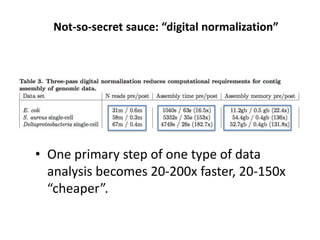
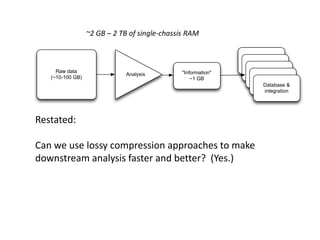
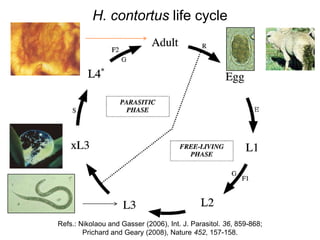
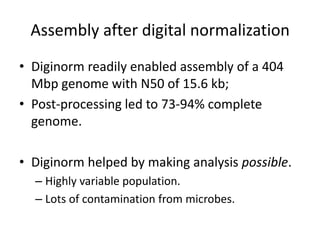
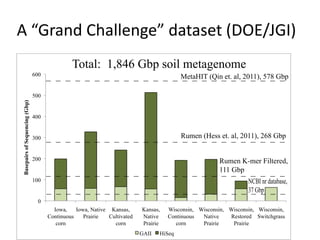
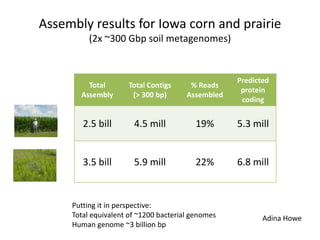
This document discusses the challenges of analyzing large genomic datasets and summarizes some of the speaker's research applying new techniques to address these challenges. The speaker develops tools to help biologists efficiently analyze vast amounts of sequencing data. One key technique is "digital normalization" which can speed up analysis by 20-200x while reducing data volume. The speaker has applied this to assemble genomes like the parasitic nematode H. contortus and soil metagenomes. The goal is to make downstream data analysis faster, better, and help answer biological questions.