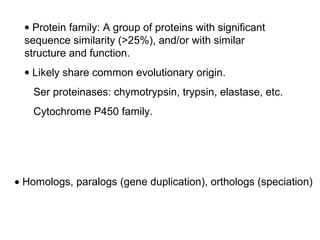
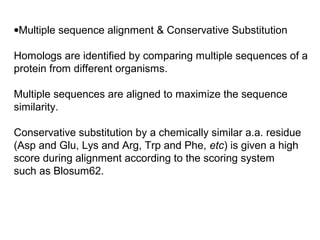
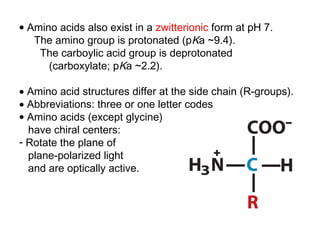
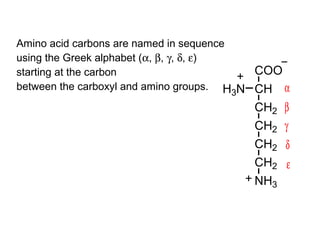
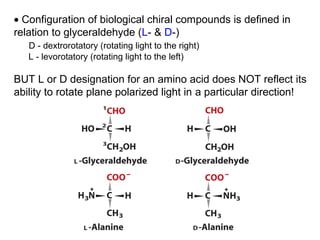
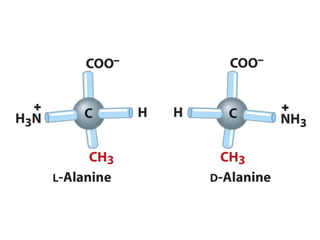
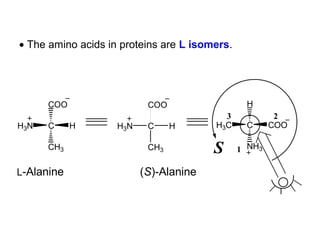
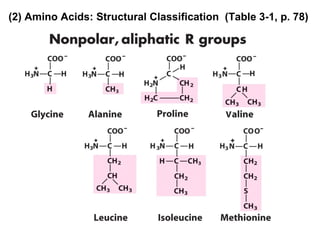
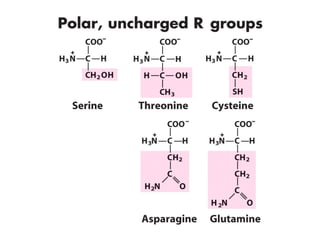
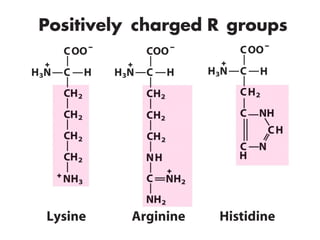
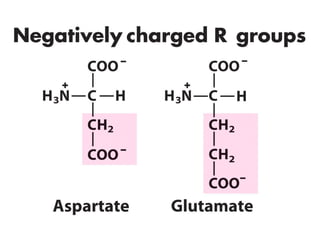
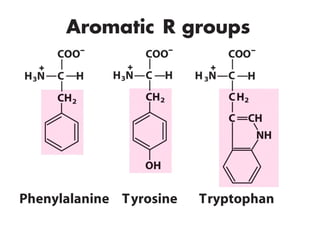
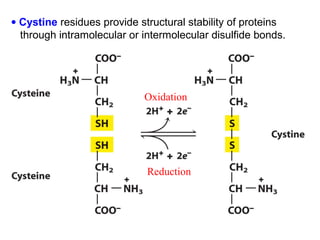
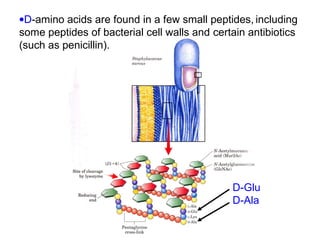
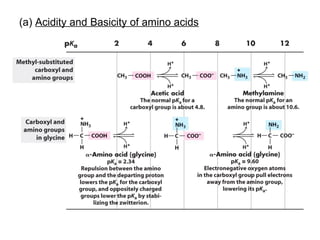
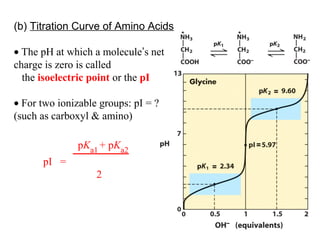
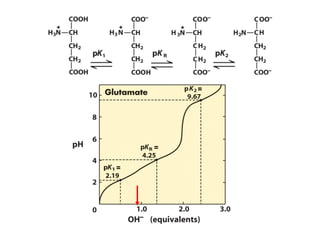
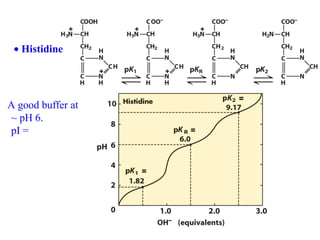
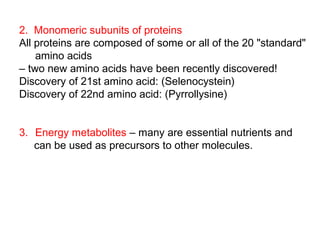
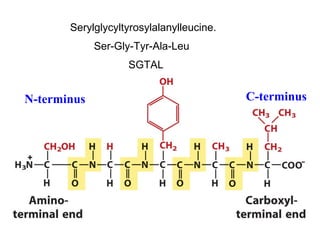
This document discusses the classification and structure of amino acids. It begins by describing the standard amino acids as α-amino acids containing an amino group and a carboxyl group. Amino acids exist in a zwitterionic form at physiological pH. They differ in their side chains and have L or D configurations. Amino acids are classified based on their side chains and functions include roles as neurotransmitters, protein subunits, and energy metabolites. Peptides and proteins are polymers of amino acids joined by peptide bonds. Protein sequences can be deduced from DNA and provide insight into structure, function, and evolution through sequence comparisons.

![[1] Amino Acids
(1) Description
• The “standard” amino acids are α-amino acids.
‒ primary amino group (−NH2)
‒ carboxylic acid group (−COOH)
•Proline is an exception
with a secondary amino group,
but, it is still referred to
as an α-amino acid.](https://image.slidesharecdn.com/08h3-130325092629-phpapp02/85/08-h3-2-320.jpg)


![[2] Peptides and Proteins (MW > 10,000):
Polymers of amino acids
Peptide synthesis:
Energetically unfavourable
(∆G > 0)
→ Couple with energetically
favourable reaction(s)
(Leaving group activation)
(Lehninger Fig 27-14, p.1052.)](https://image.slidesharecdn.com/08h3-130325092629-phpapp02/85/08-h3-22-320.jpg)

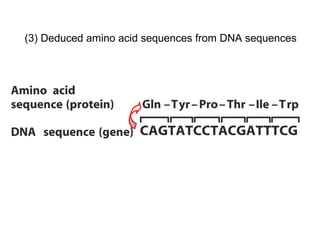

![[4] Protein Sequences and Evolution
• Genes and proteins from closely related organisms
should be similar.
• The sequences increasingly diverge as the evolutionary
distance between two organisms increases.
• Conserved a.a. residues: amino acid residues essential
for function and structure are conserved throughout the
evolution.
• Variable residues: Those less important vary over time.
⇒ polymorphism](https://image.slidesharecdn.com/08h3-130325092629-phpapp02/85/08-h3-27-320.jpg)