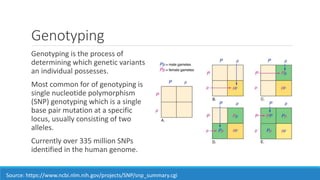
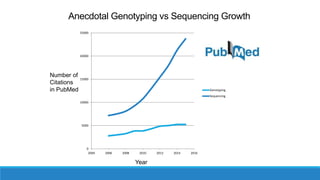
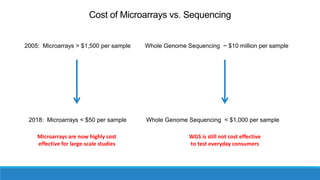
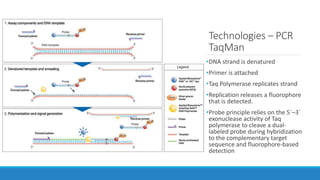
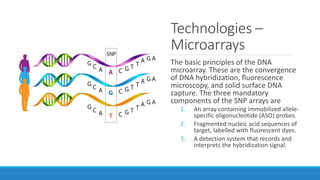
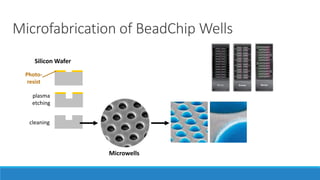
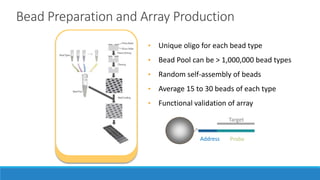
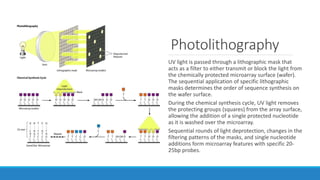
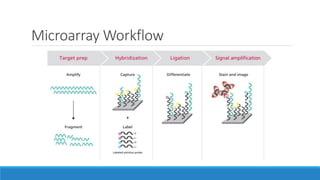
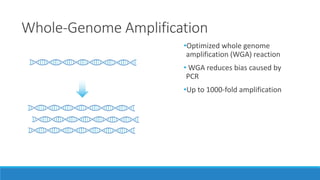
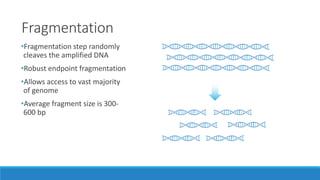
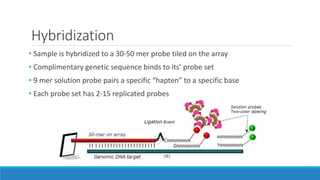
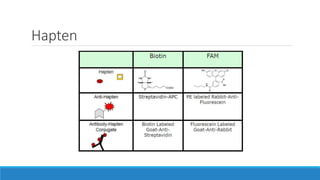
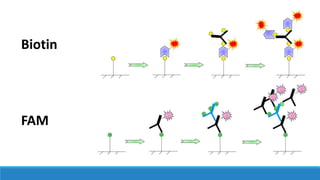
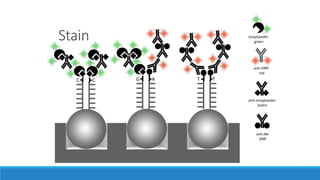
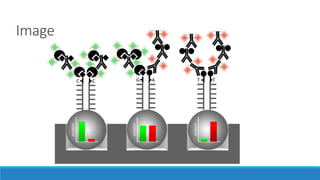
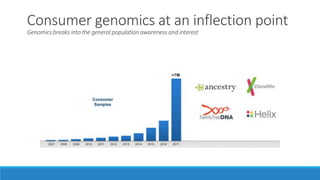
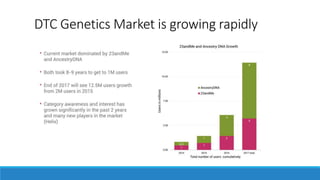
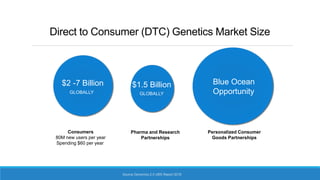
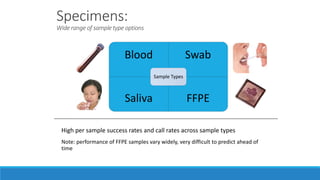
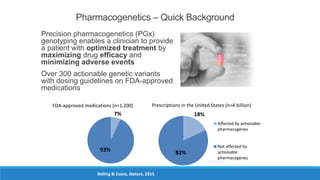
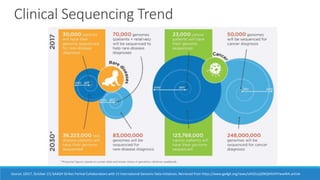
This document discusses clinical and consumer applications of microarrays and genotyping technologies. It provides an overview of genotyping and different technologies like PCR microarrays and SNP microarrays. It describes how microarrays are still useful despite the rise of sequencing due to their low cost, high throughput, and ability to test millions of markers. The document outlines several applications of microarrays like direct-to-consumer testing, pharmacogenetics, and clinical sequencing. It also discusses challenges and trends in these areas like global initiatives to increase genomic data sharing.