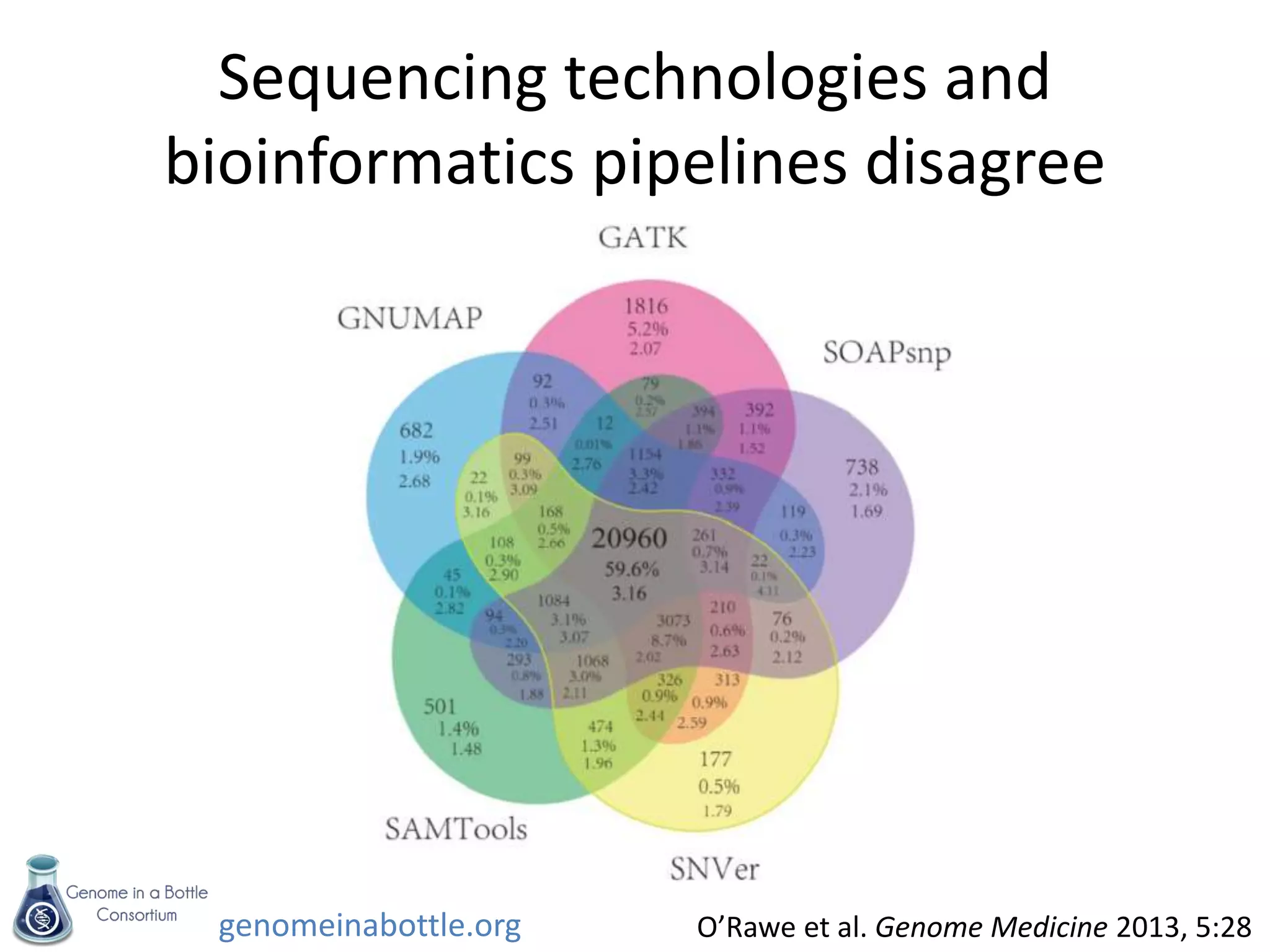
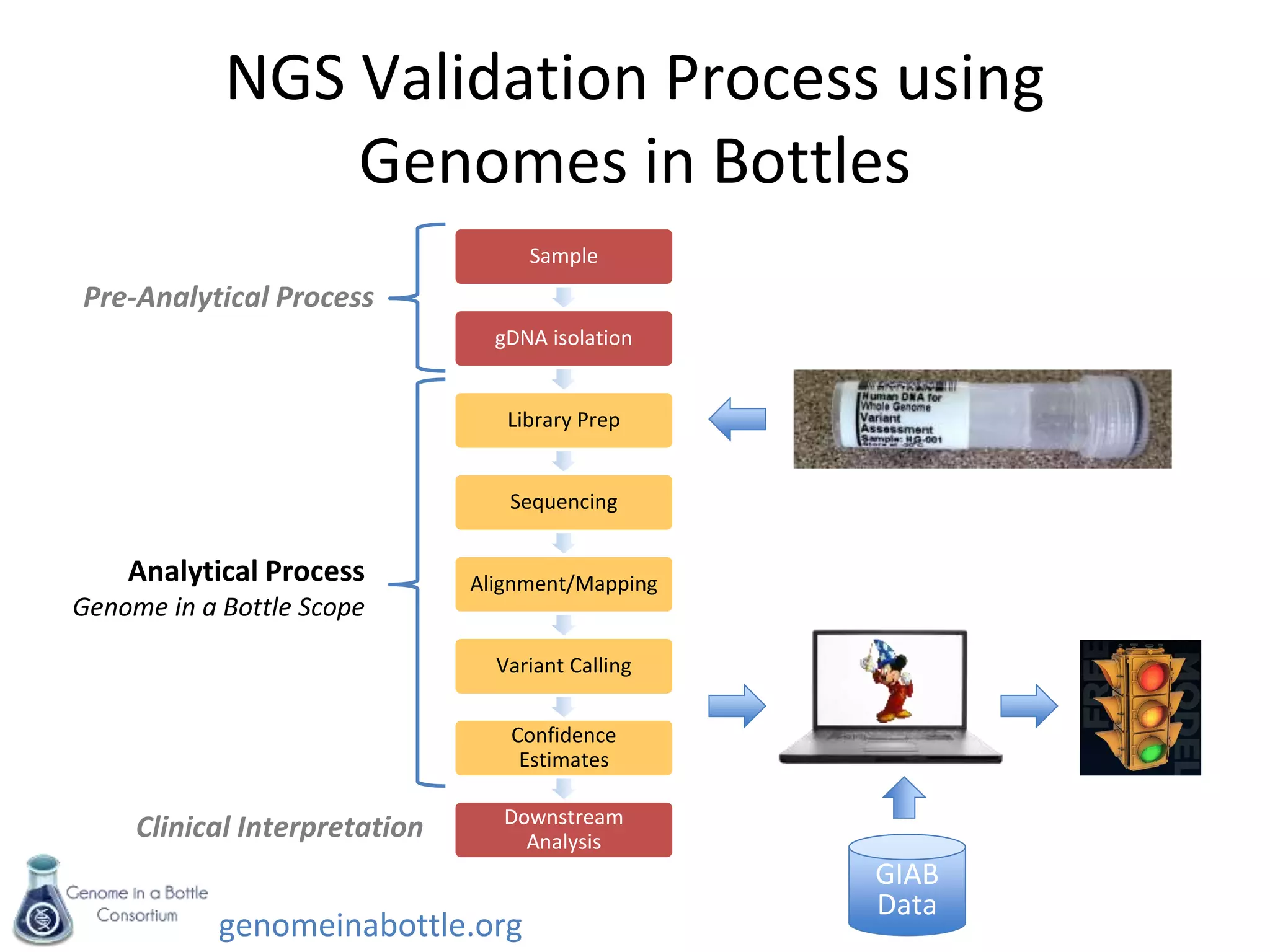
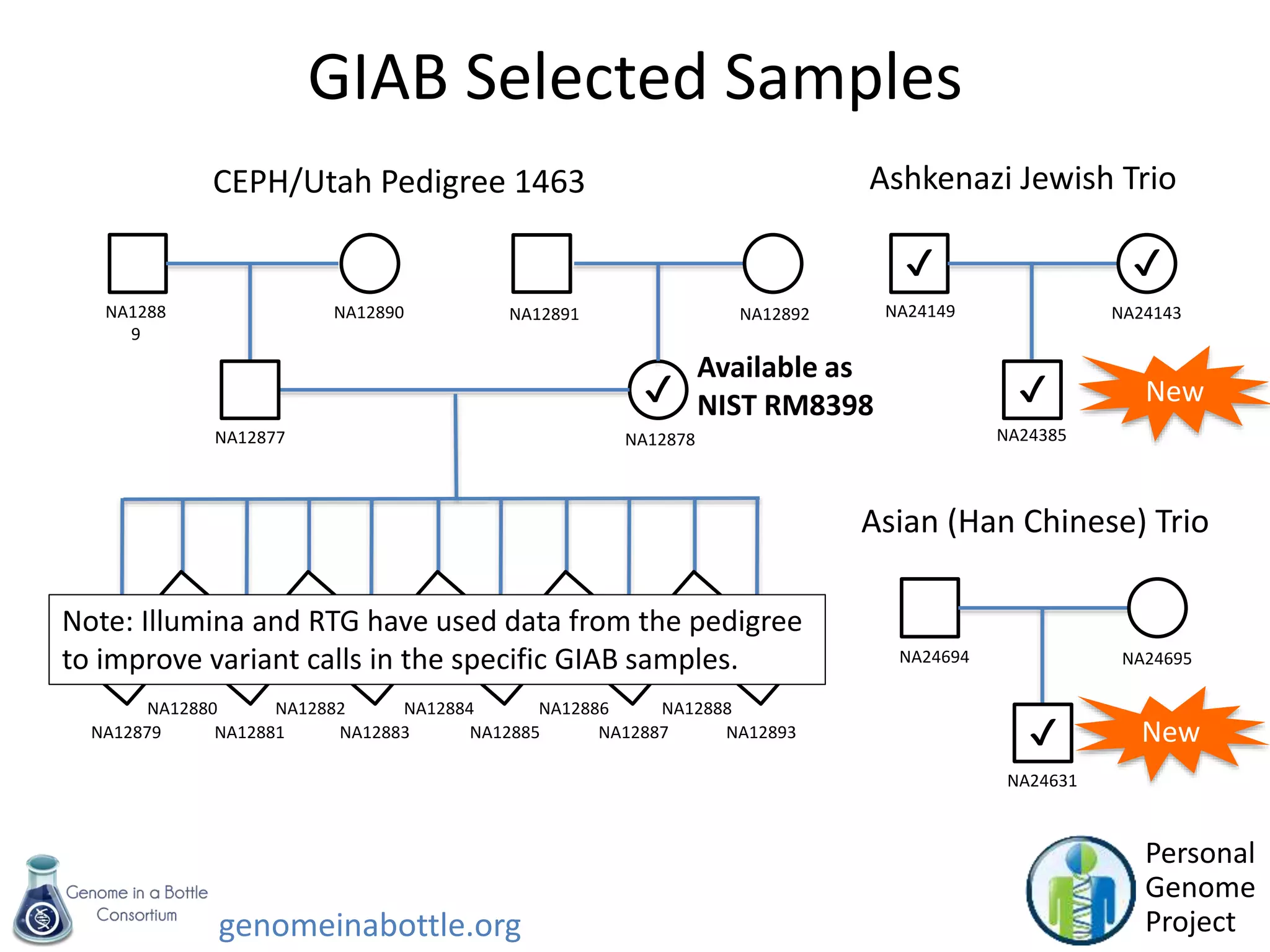
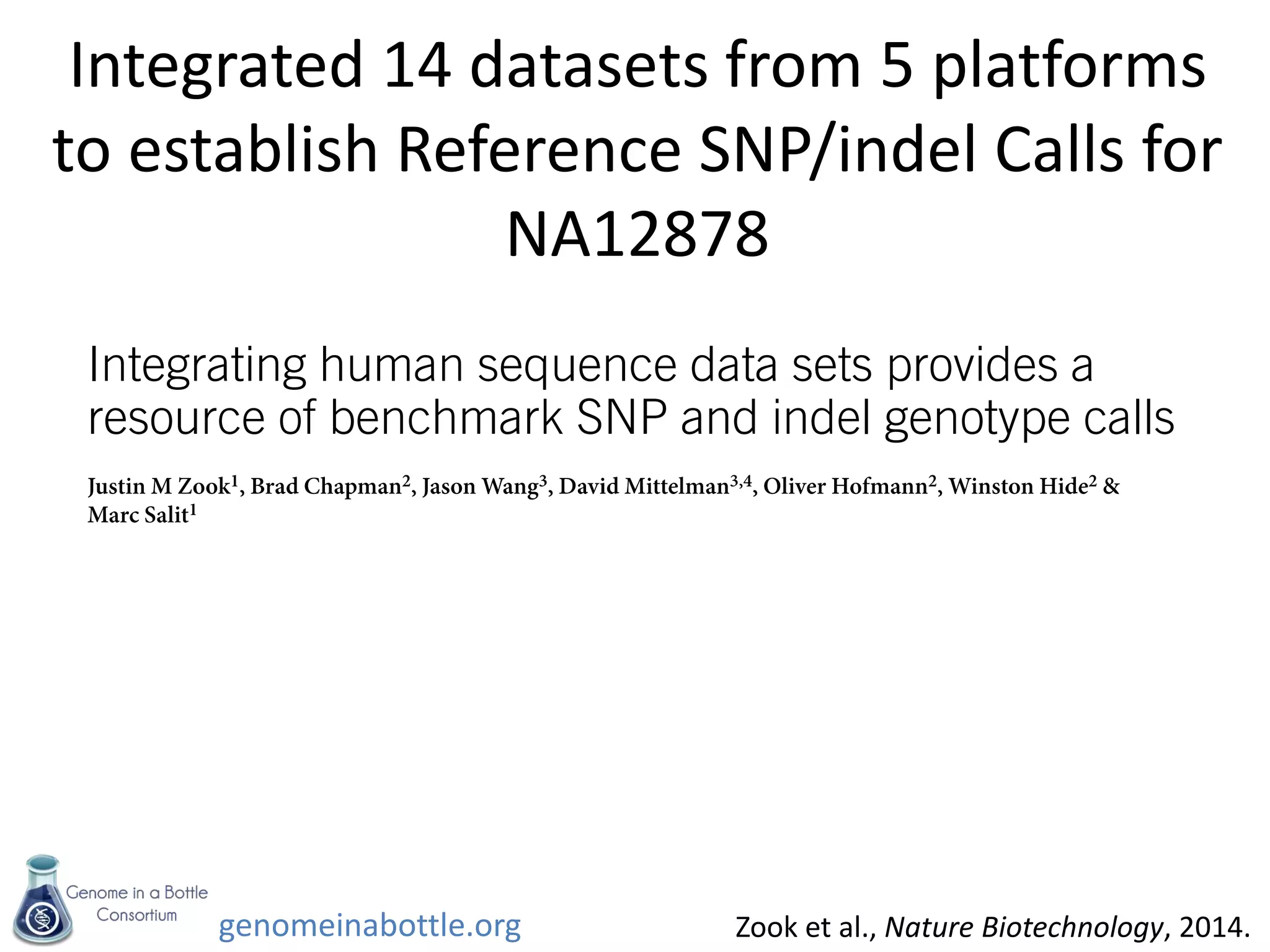
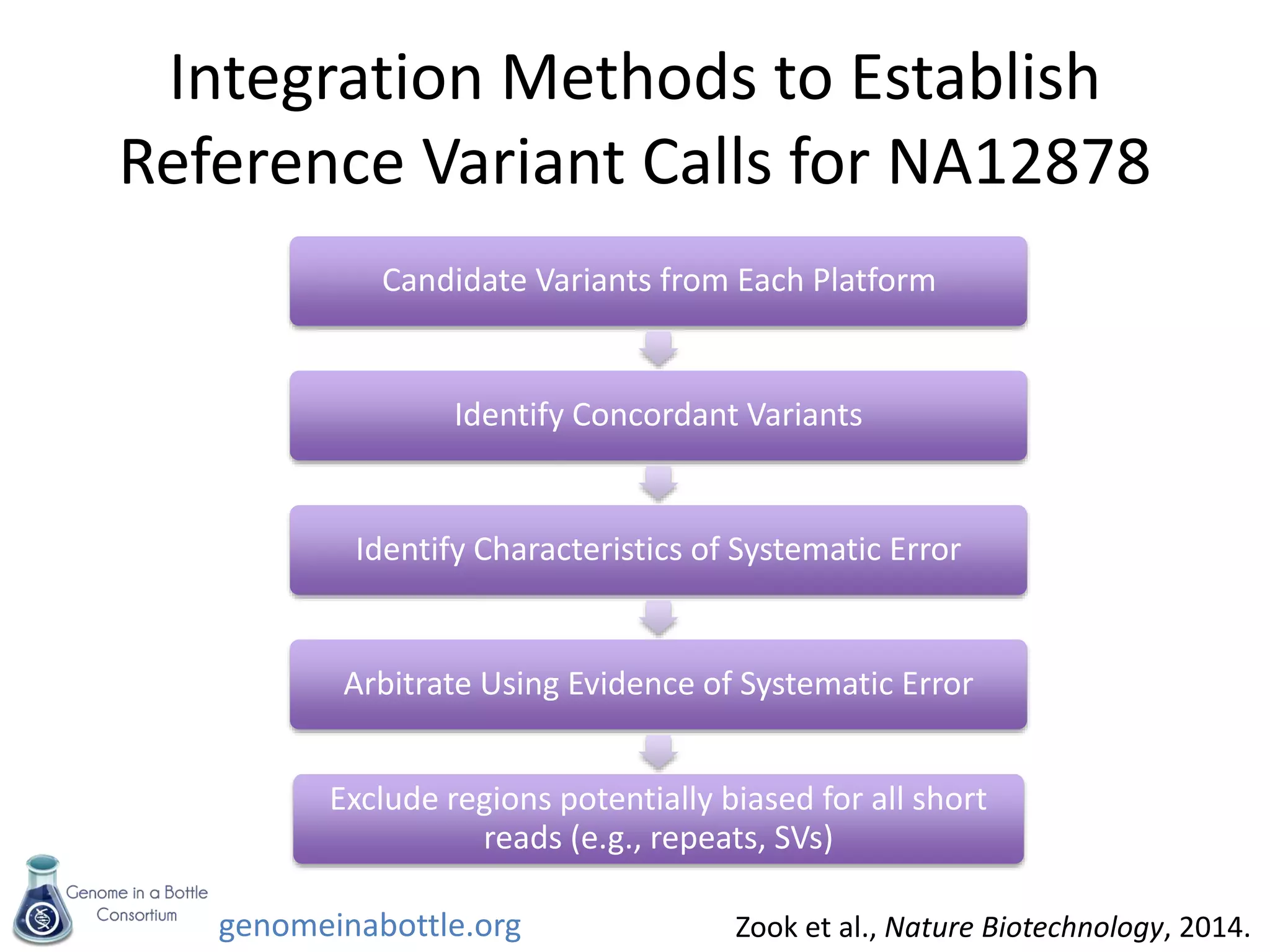
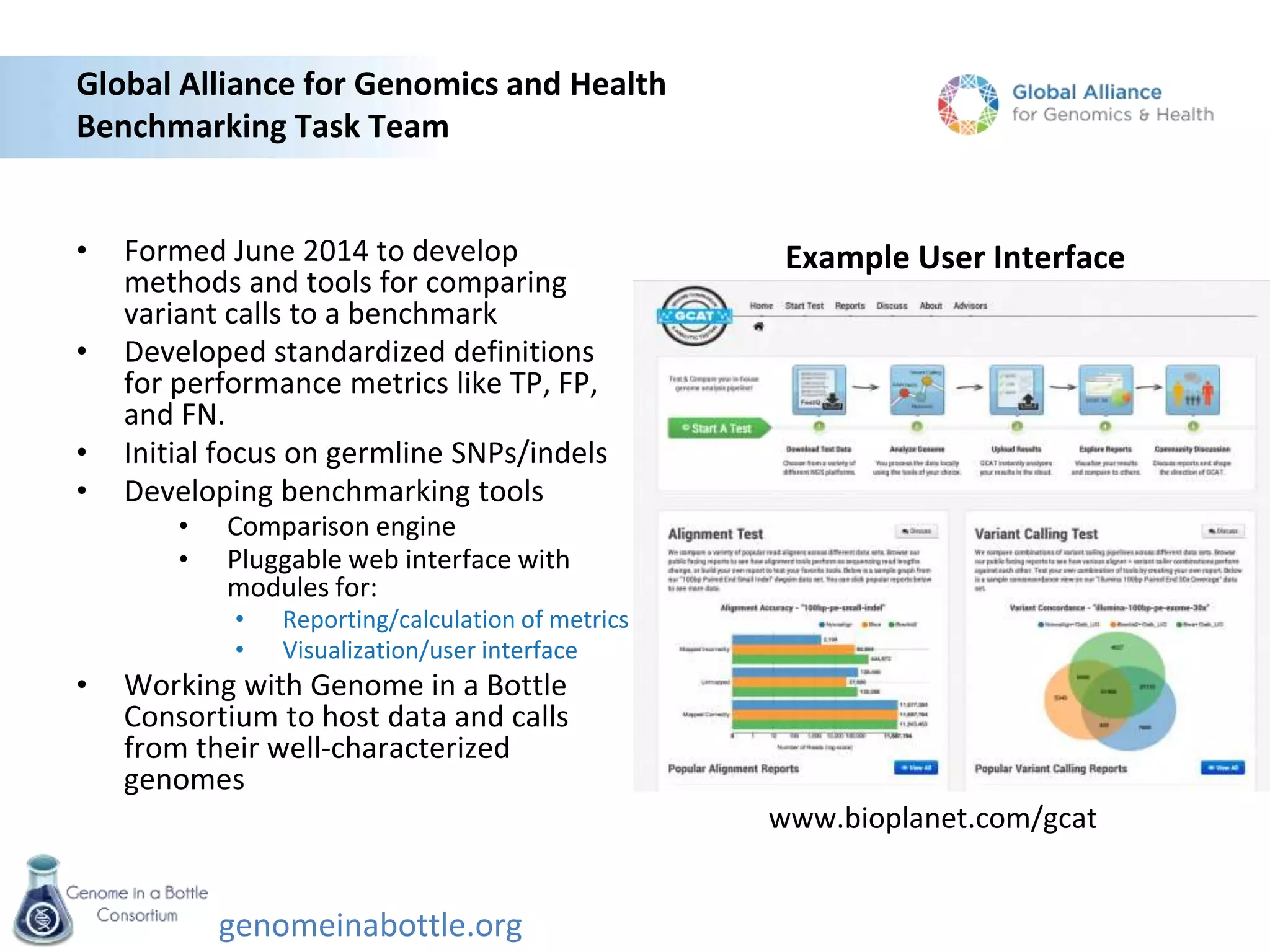
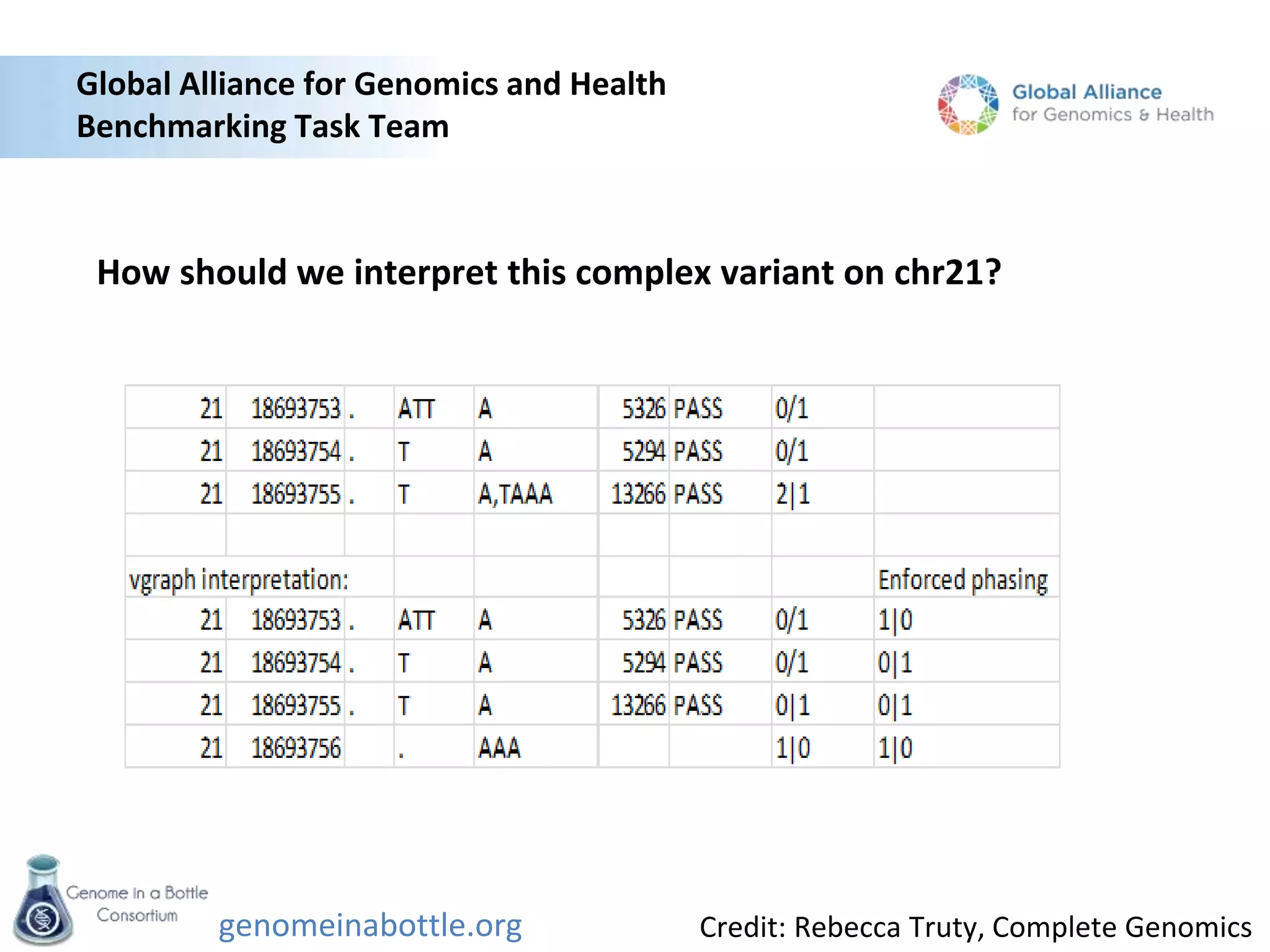
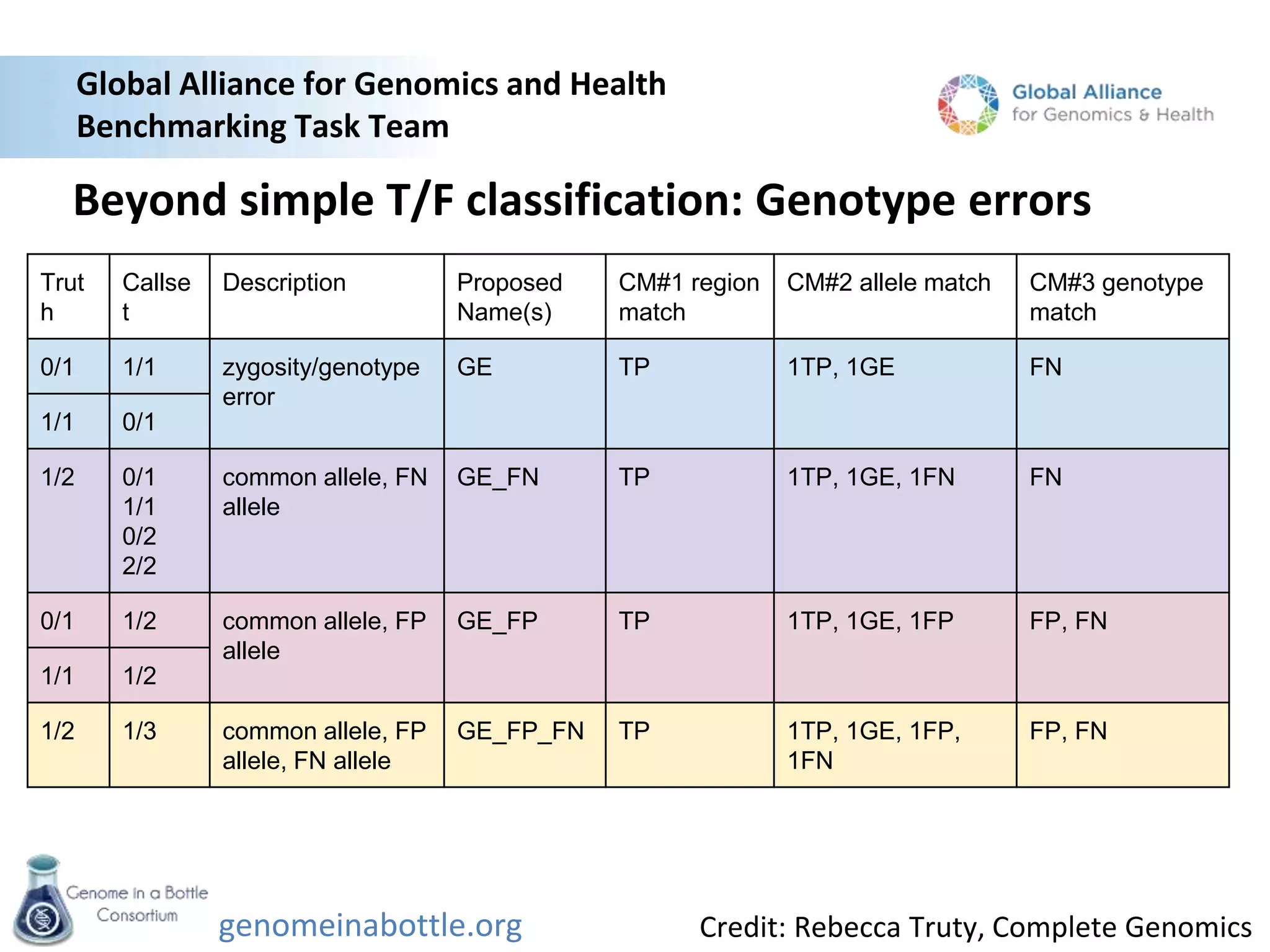
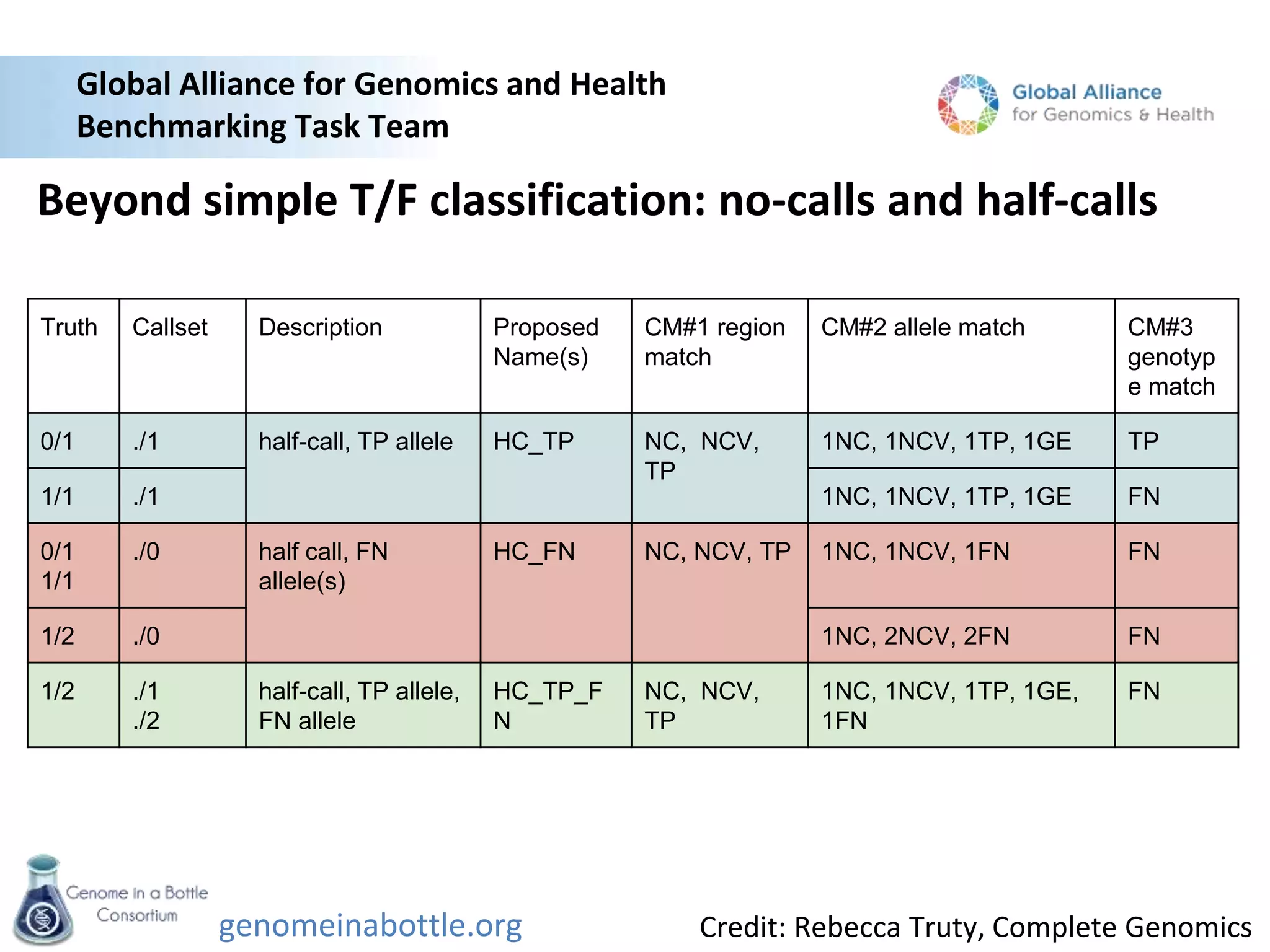
The document discusses the Genome in a Bottle Consortium's efforts to generate reference materials and data to evaluate the accuracy of human genome sequencing and variant calling. Specifically:
- The consortium is developing reference samples with well-characterized genomes to test sequencing platforms and bioinformatics methods.
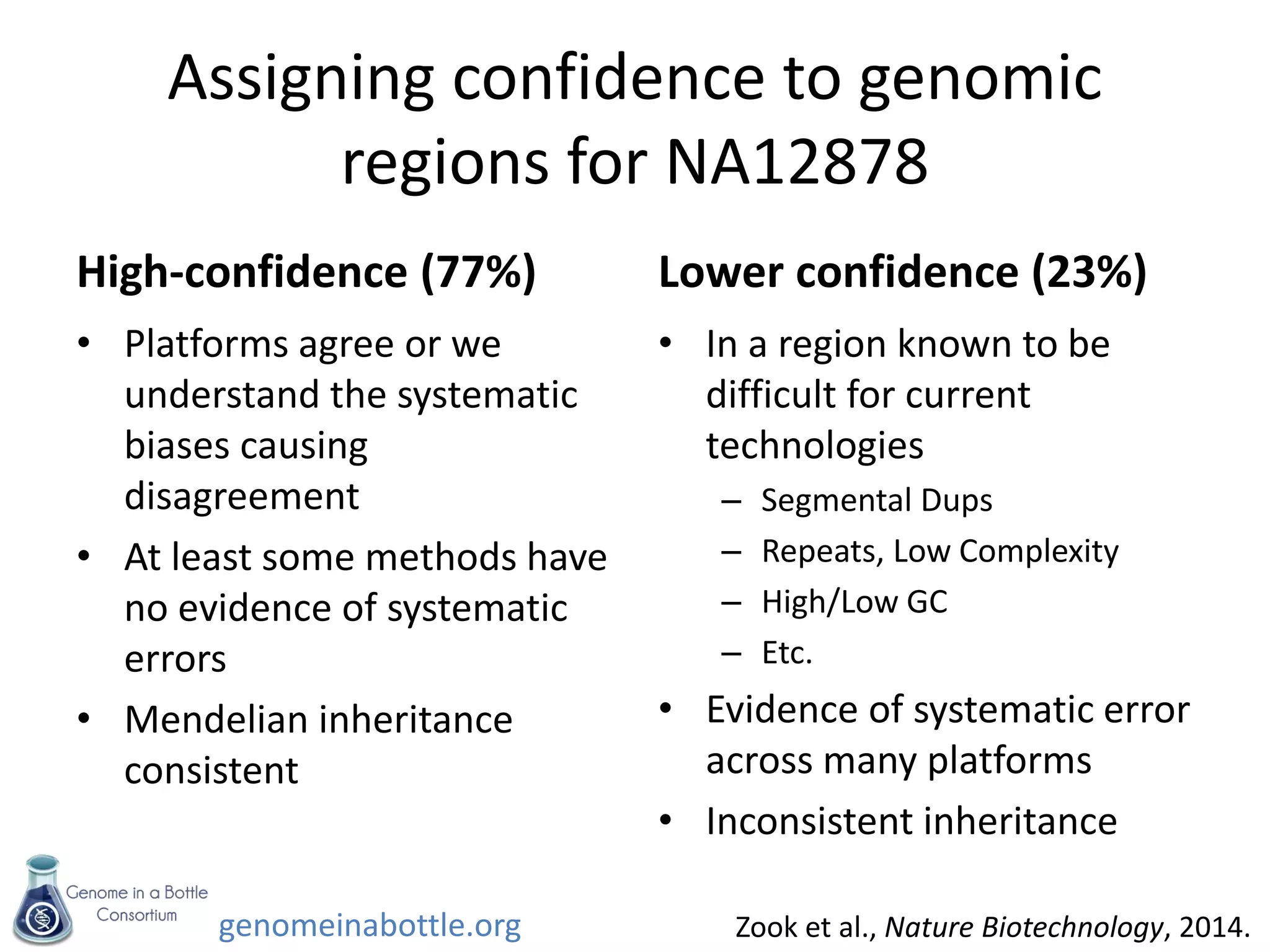
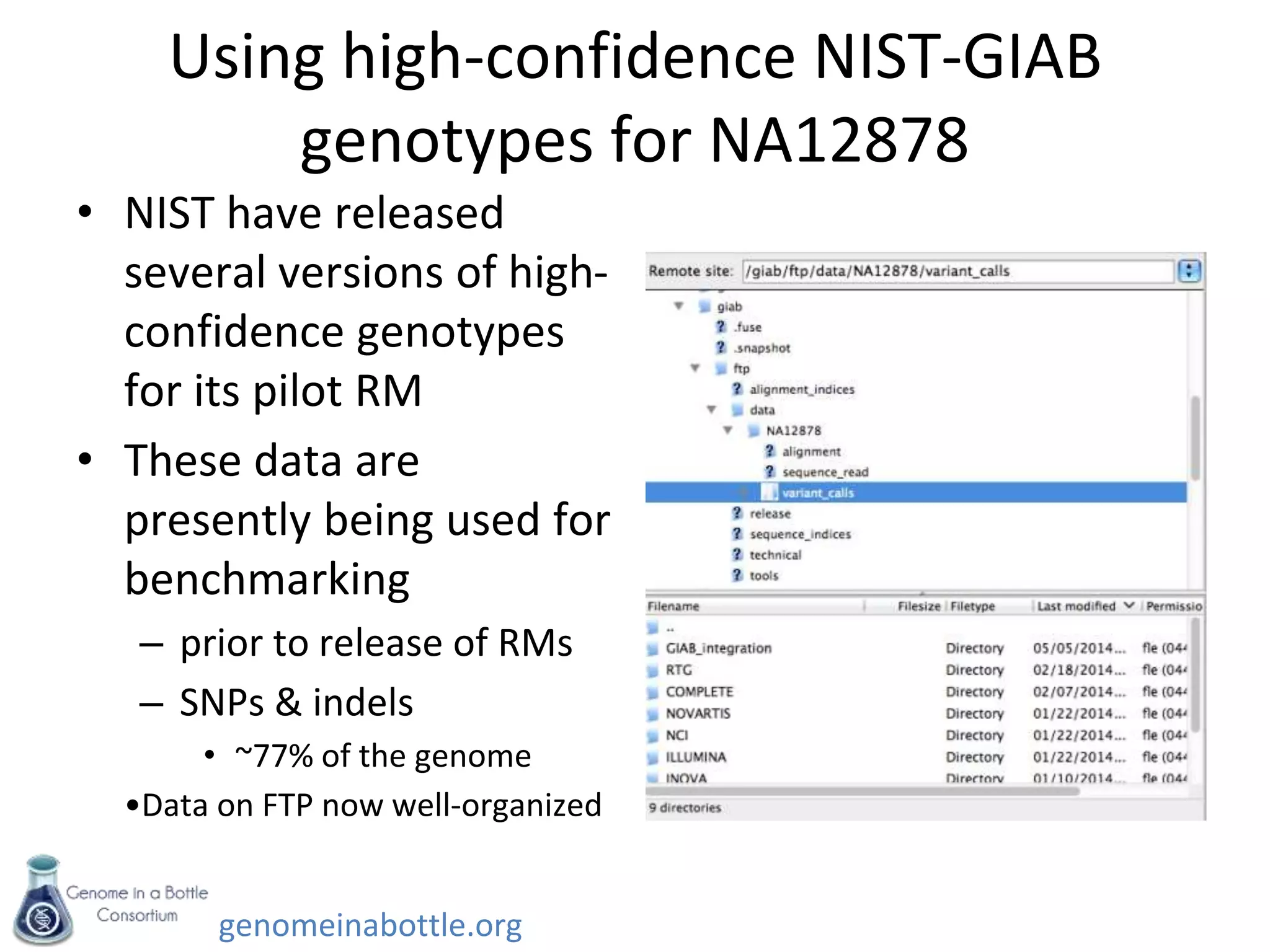
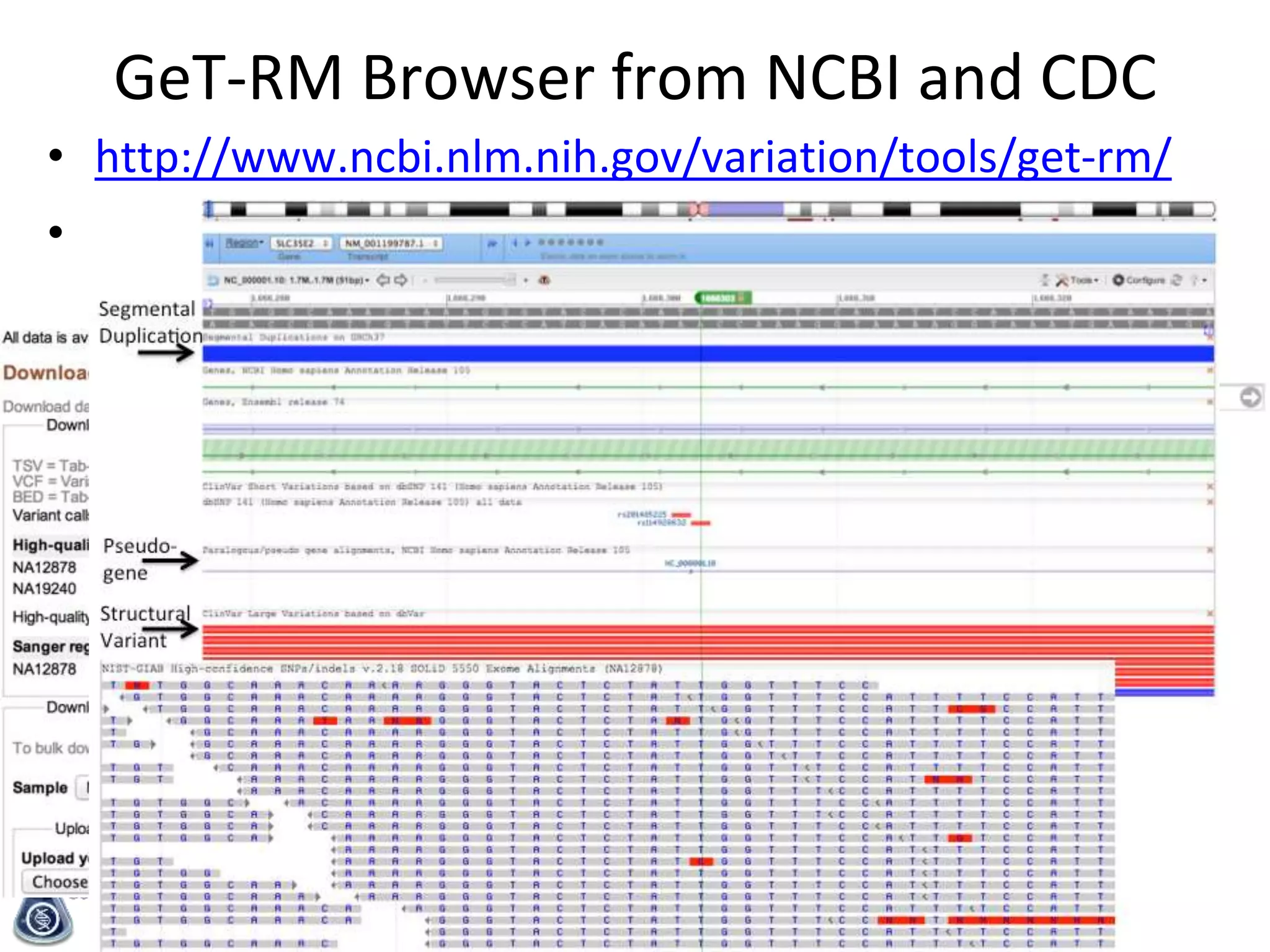
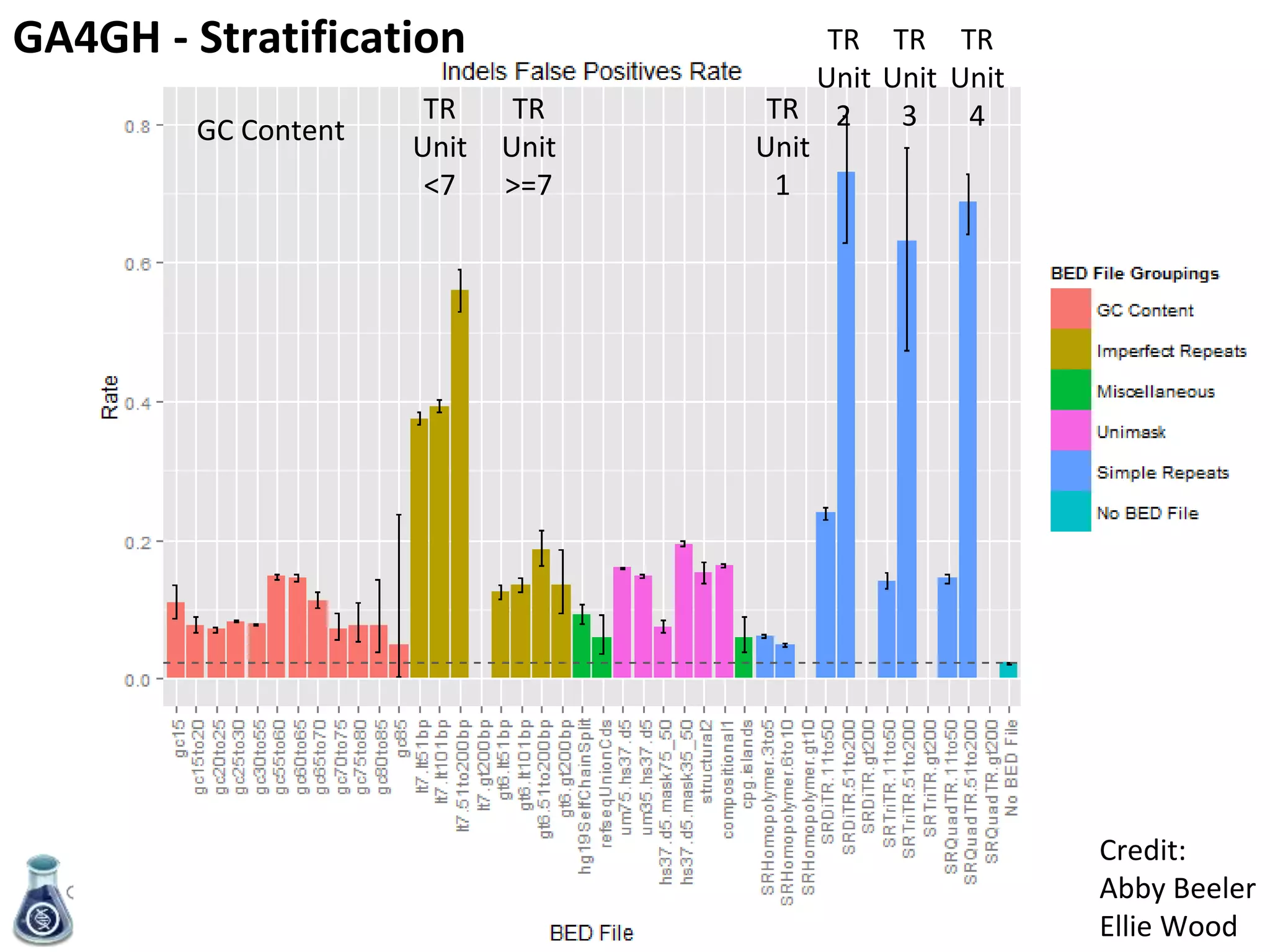
- An initial sample, NA12878, has been extensively sequenced and analyzed to generate high-confidence variant calls across 77% of the genome.
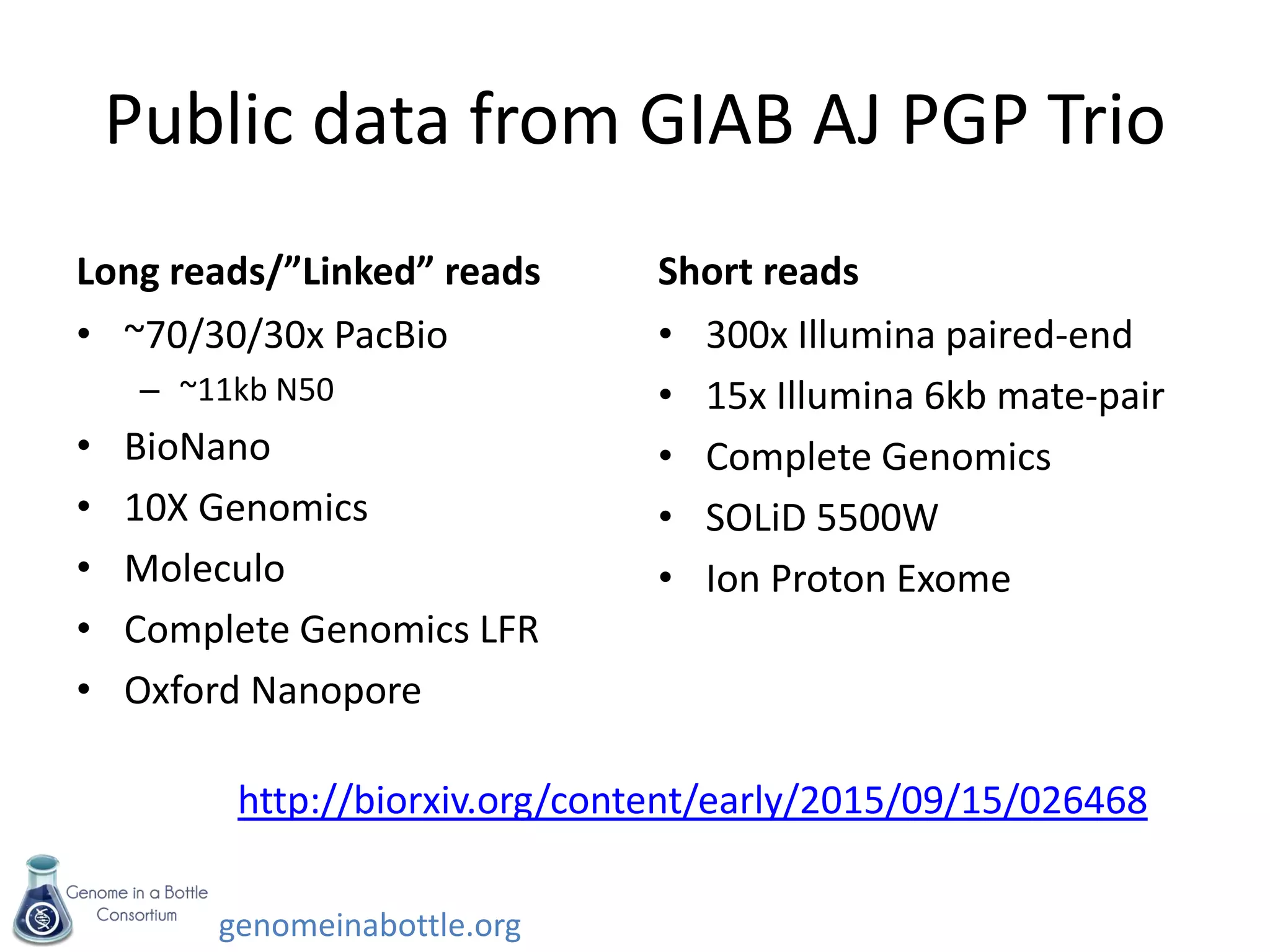
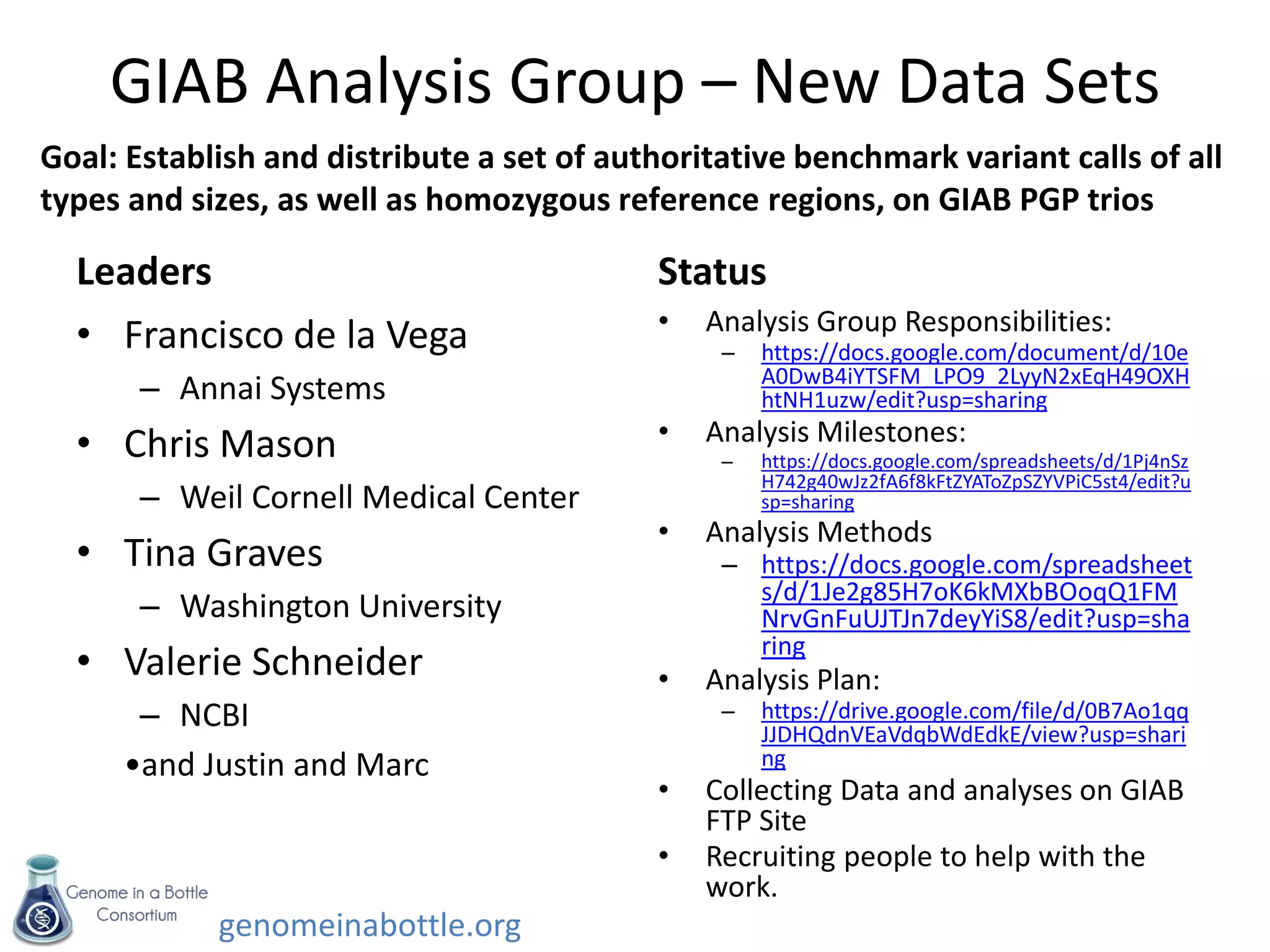
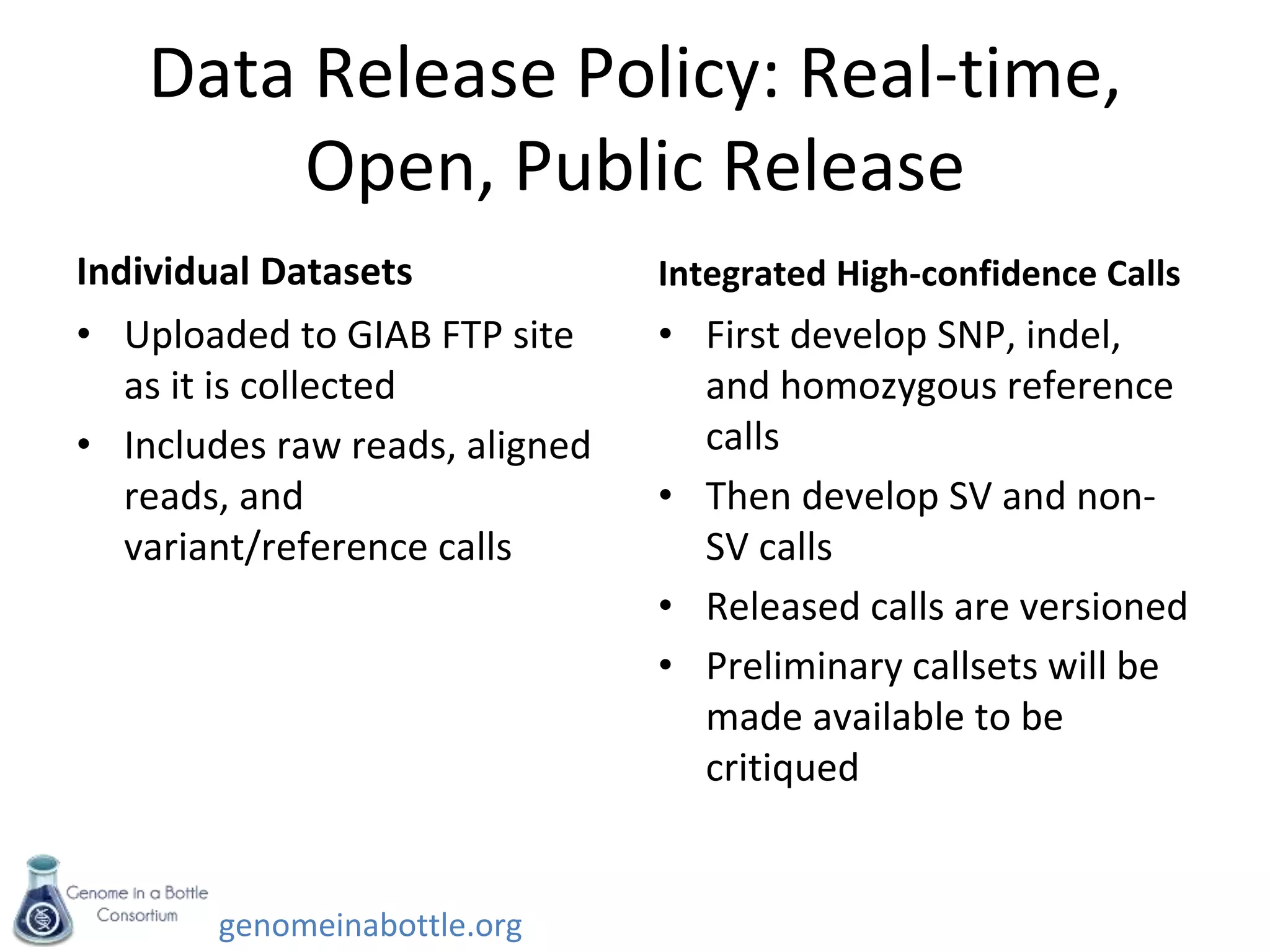
- Efforts are ongoing to expand the reference data to additional samples from different populations and integrate data from multiple sequencing technologies.
- The goal is to enable standardized evaluation, benchmarking and regulatory oversight of clinical genome sequencing.