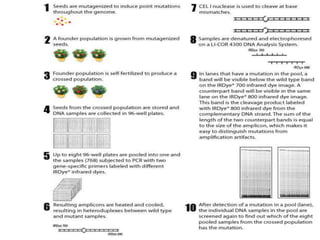
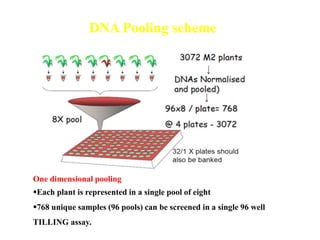
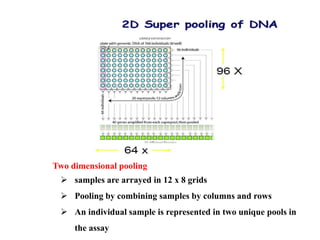
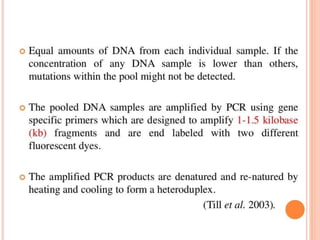
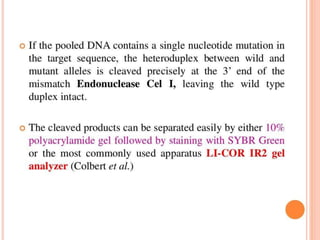
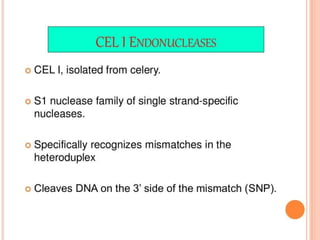
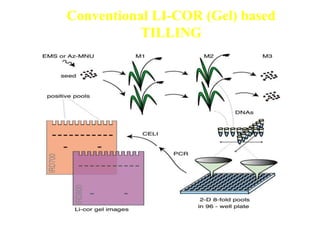
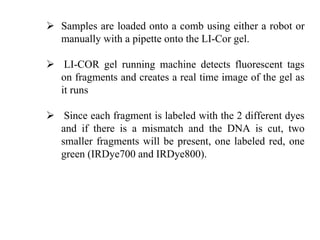
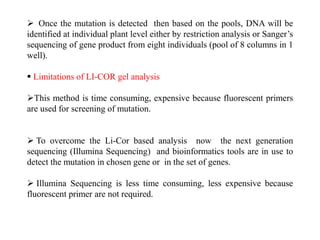
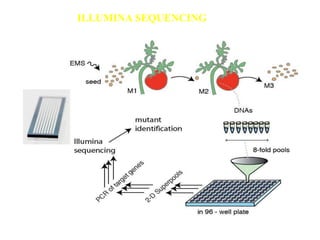
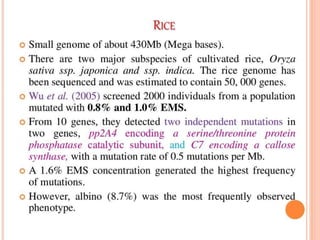
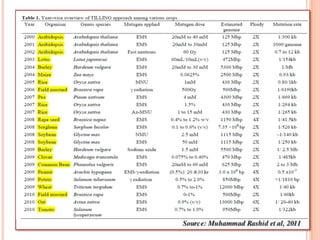
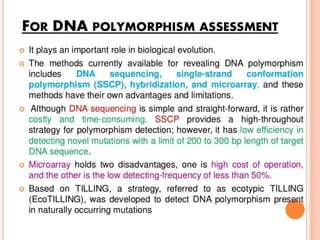
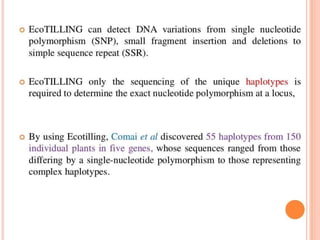
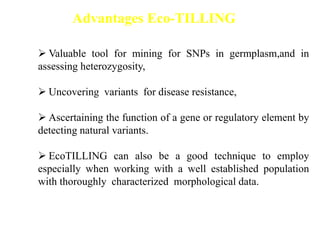
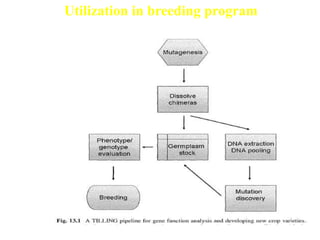
This document discusses TILLING and Eco-TILLING techniques for crop improvement. It describes the limitations of existing gene knockout methods and outlines the TILLING workflow, including DNA pooling schemes and both conventional LI-COR gel-based and newer Illumina sequencing-based methods for detecting mutations. Eco-TILLING can be used to mine germplasm for SNPs and assess heterozygosity to uncover variants related to disease resistance or gene function. Both TILLING and Eco-TILLING can help characterize natural variation and are useful techniques in plant breeding programs.