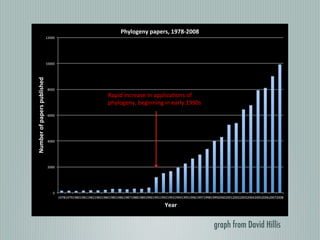
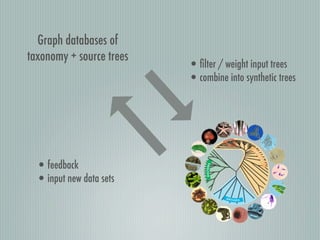
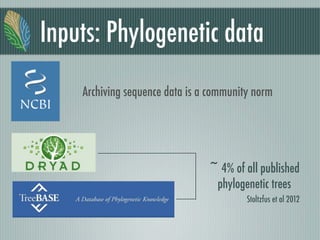
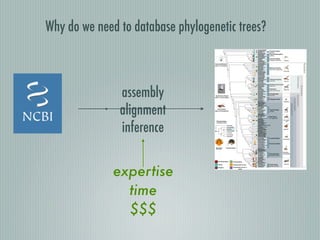
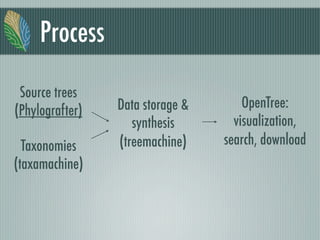
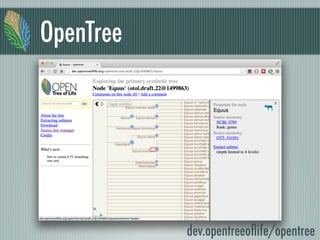
This document summarizes the goals and progress of the Open Tree of Life project, which aims to synthesize a complete draft tree of life using existing phylogenetic data. The project has collected phylogenetic data from over 7000 studies and stored it using graph databases. An open public interface allows users to browse, download and query the tree. The project is on track to release an initial draft tree in year 1 and refine it based on user feedback in year 2, while expanding collaborations and incentives for data contributors.