This document discusses antibiotic susceptibility testing for aerobic bacteria. It describes various antibiotic classes and provides examples of antibiotics within each class. It also discusses new antibiotics for treating multi-drug resistant gram-negative bacteria. The major mechanisms of antibiotic resistance are outlined. Finally, it summarizes methods for susceptibility testing and discusses antibiotic resistance in important bacteria such as MRSA, VRE, ESBL producers, and carbapenemase producers.
















![Extended Spectrum Beta Lactamase [ESBL]
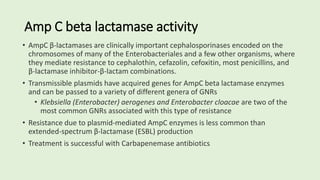
•Enzymes produced by Enteric gram-negative bacilli
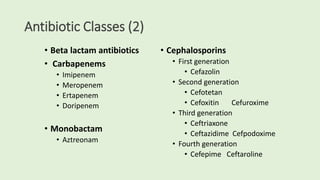
• Beta lactamase enzymes confer resistance to Cephalosporins, Penicillins
and Monobactam (Aztreonam) antibiotics by opening the beta lactam
ring and inactivating the antibiotic
• ESBLs do not attack Cephamycin (cefoxitin, cefotetan) or the Carbapenem
antibiotic classes
• Most common in Escherichia coli, Klebsiella spp and Proteus mirabilis
•Plasmid mediated CTX-M beta lactamases are the most common ESBL
enzymes produced in the US currently, but many more ESBL types can be
found worldwide
•Therapy for ESBL producing gram-negative rods include the Carbapenems:
Imipenem, Meropenem, Doripenem, and Ertapenem](https://image.slidesharecdn.com/webpagesusceptibility2023-230218020639-54abb55a/85/Susceptibility-2023-23-320.jpg)