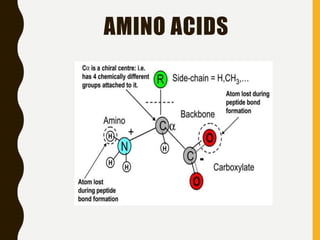
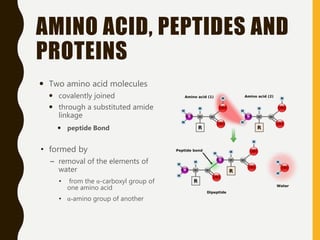
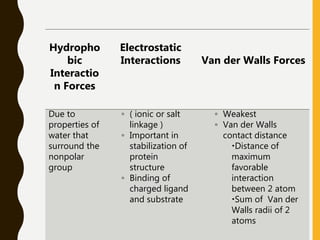
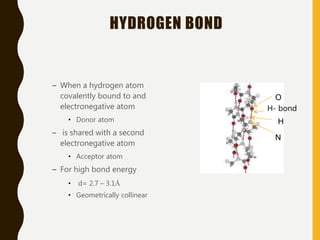
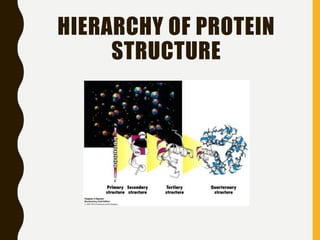
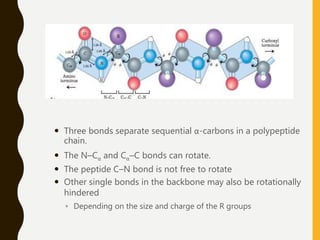
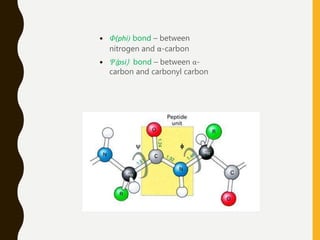
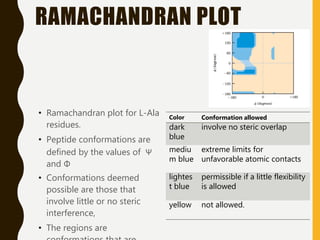
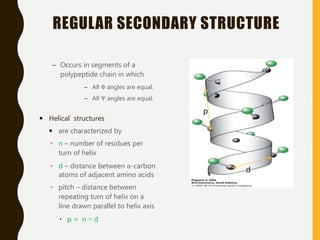
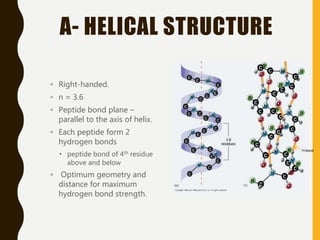
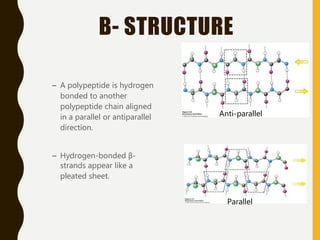
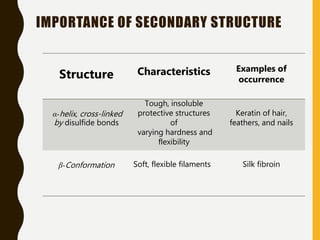
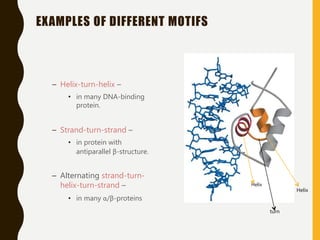
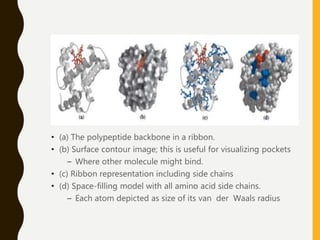
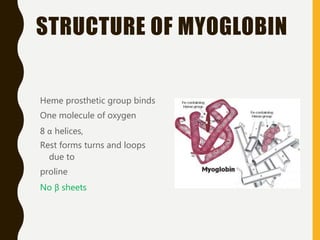
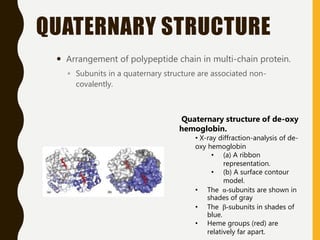
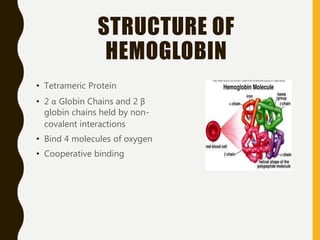
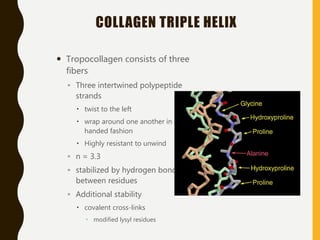
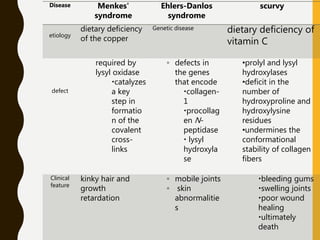
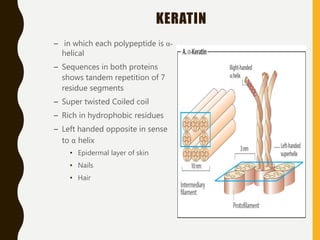
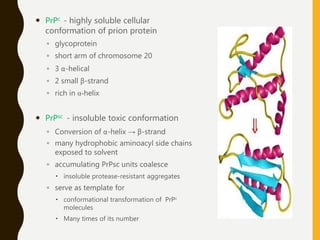
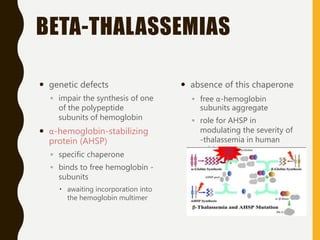
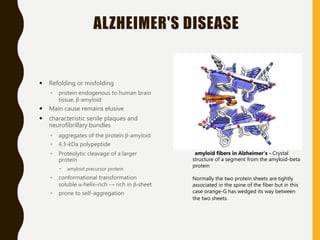
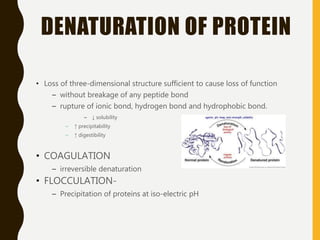
The document provides an extensive overview of protein structure, detailing primary, secondary, tertiary, and quaternary structures, as well as the importance of amino acids, peptide bonds, and protein folding. It discusses the roles of various forces, such as non-covalent interactions, in stabilizing protein conformations and links protein structure to diseases and clinical correlations. Additionally, it touches on topics like unstructured proteins, protein complexes, and the implications of protein misfolding in diseases such as Alzheimer's and prion diseases.