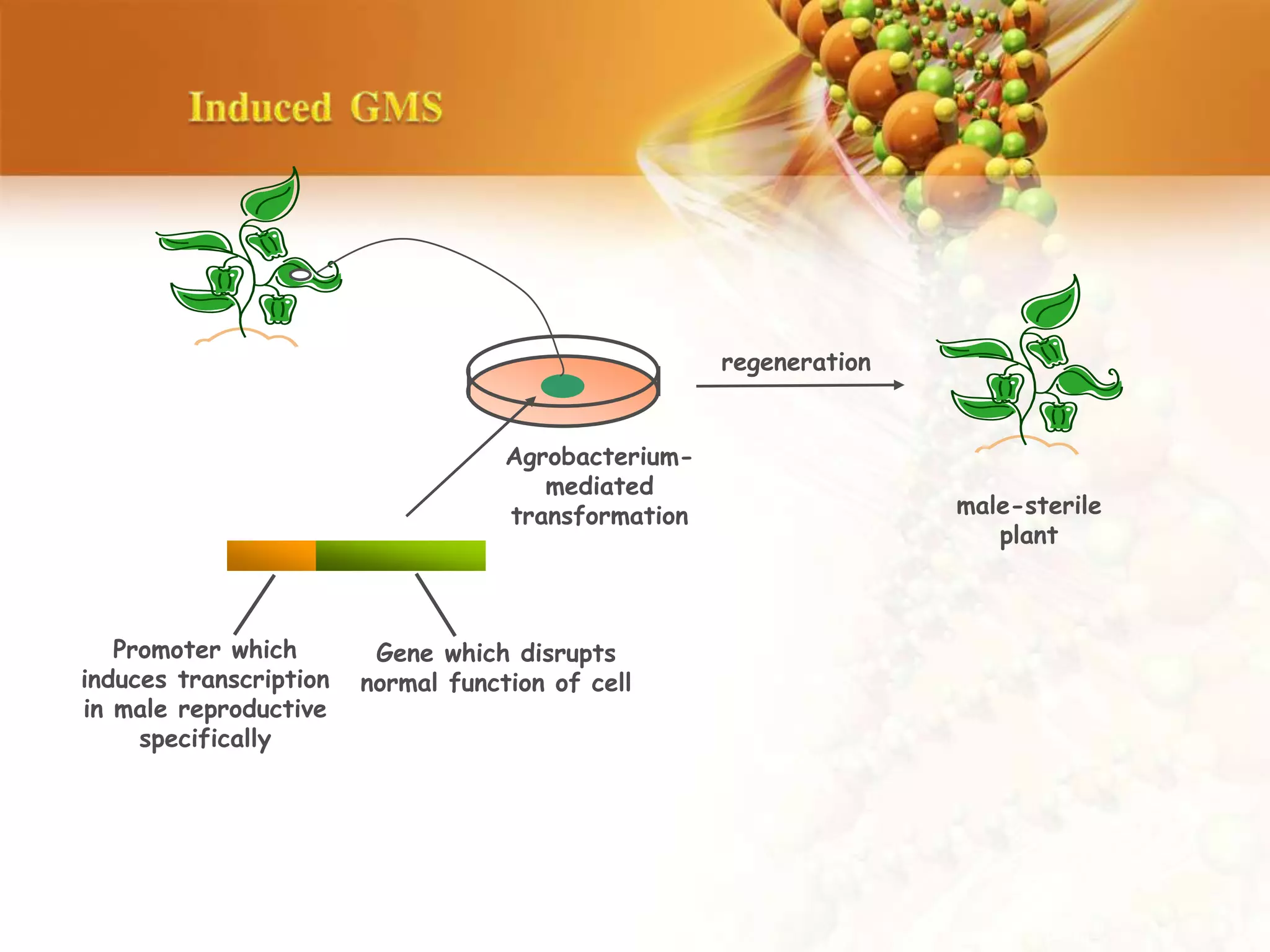
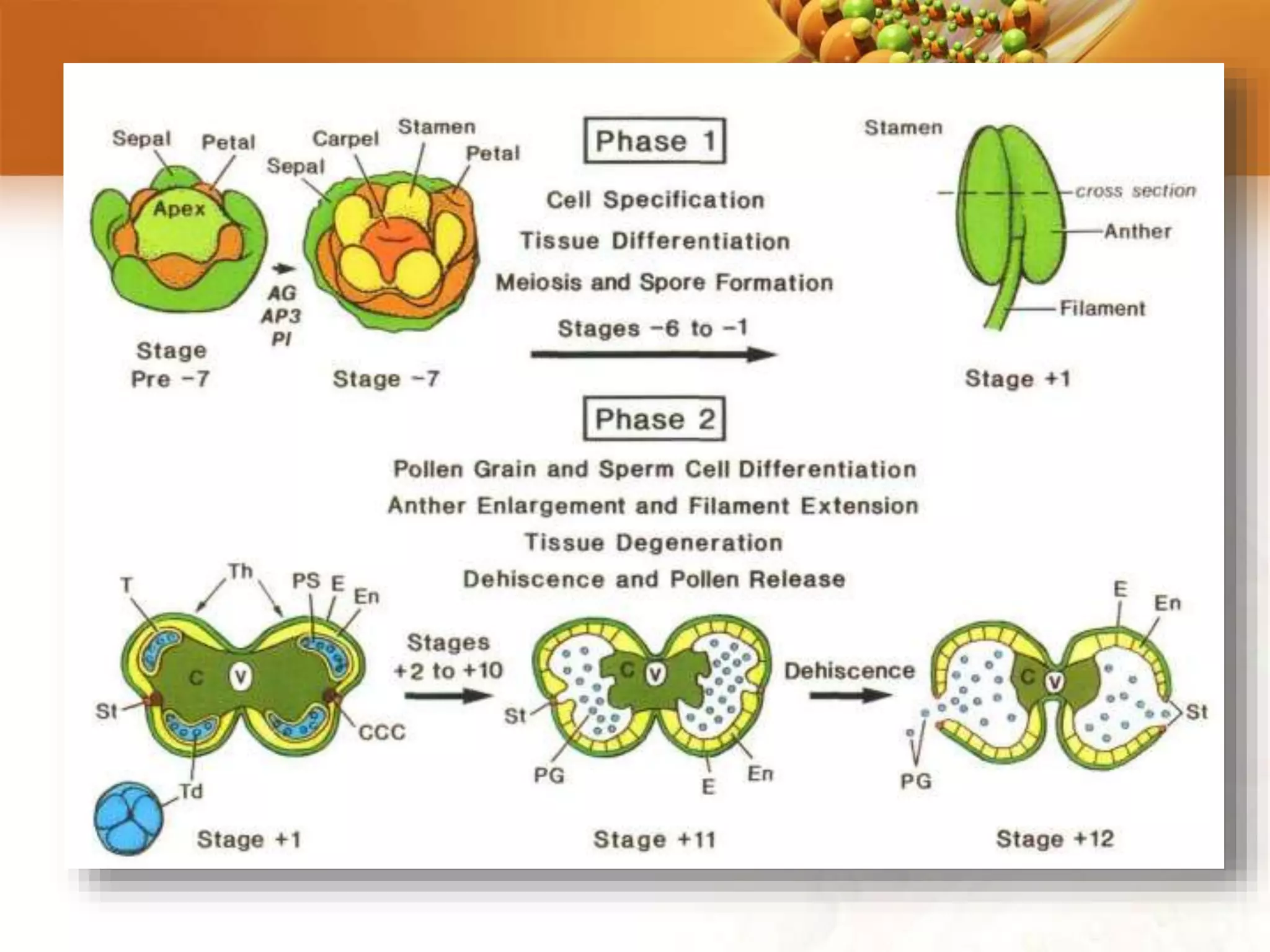
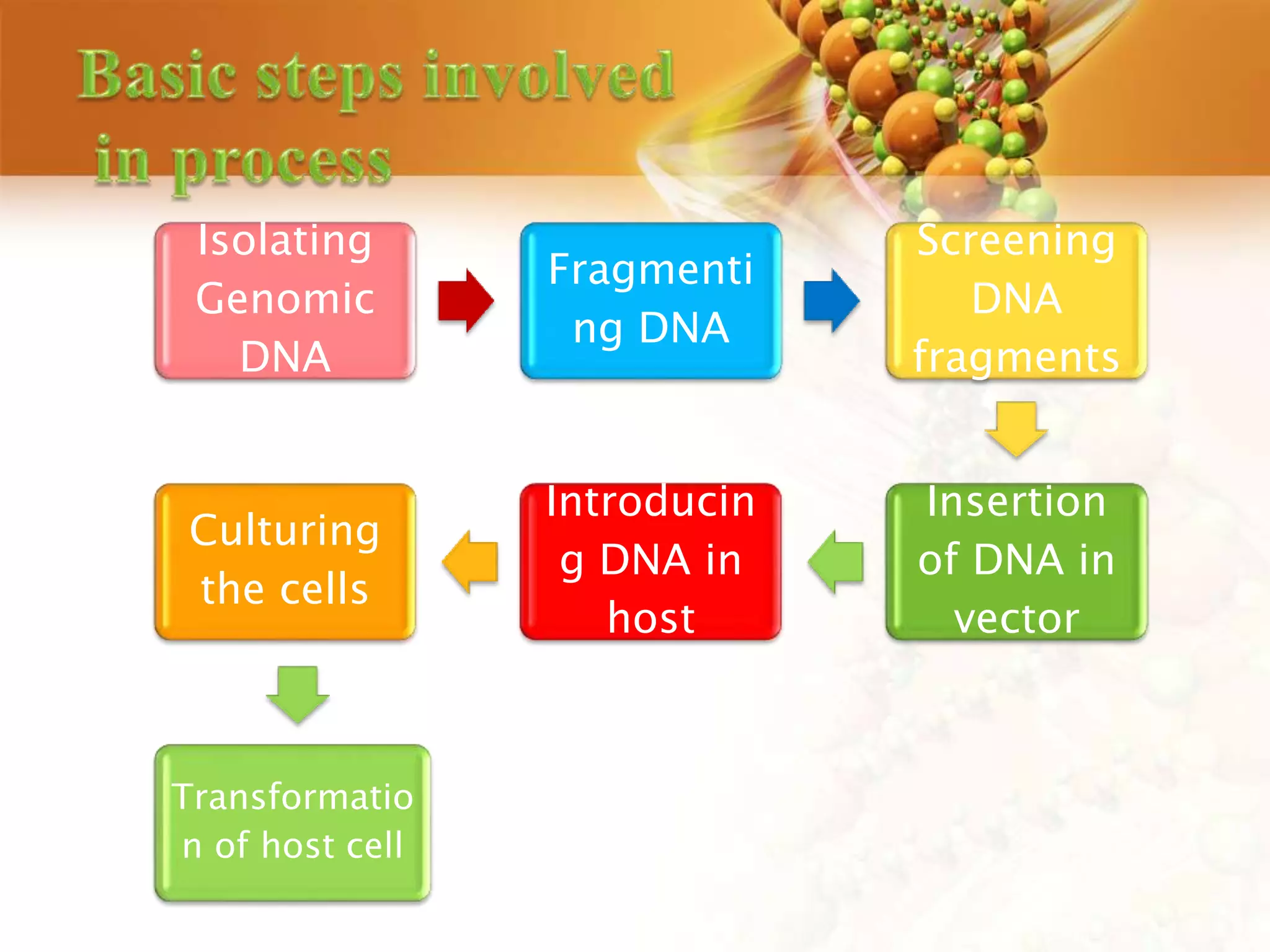
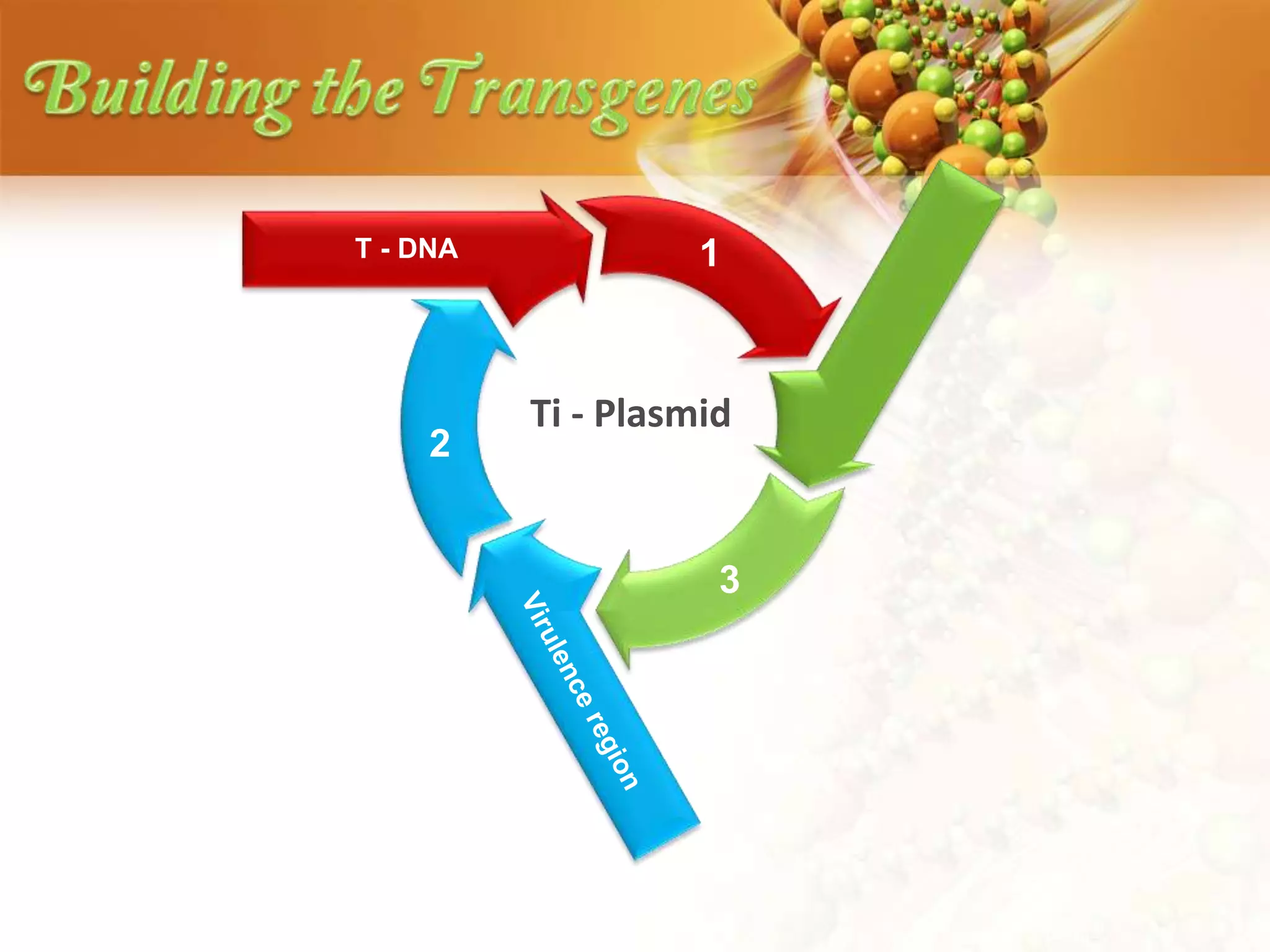
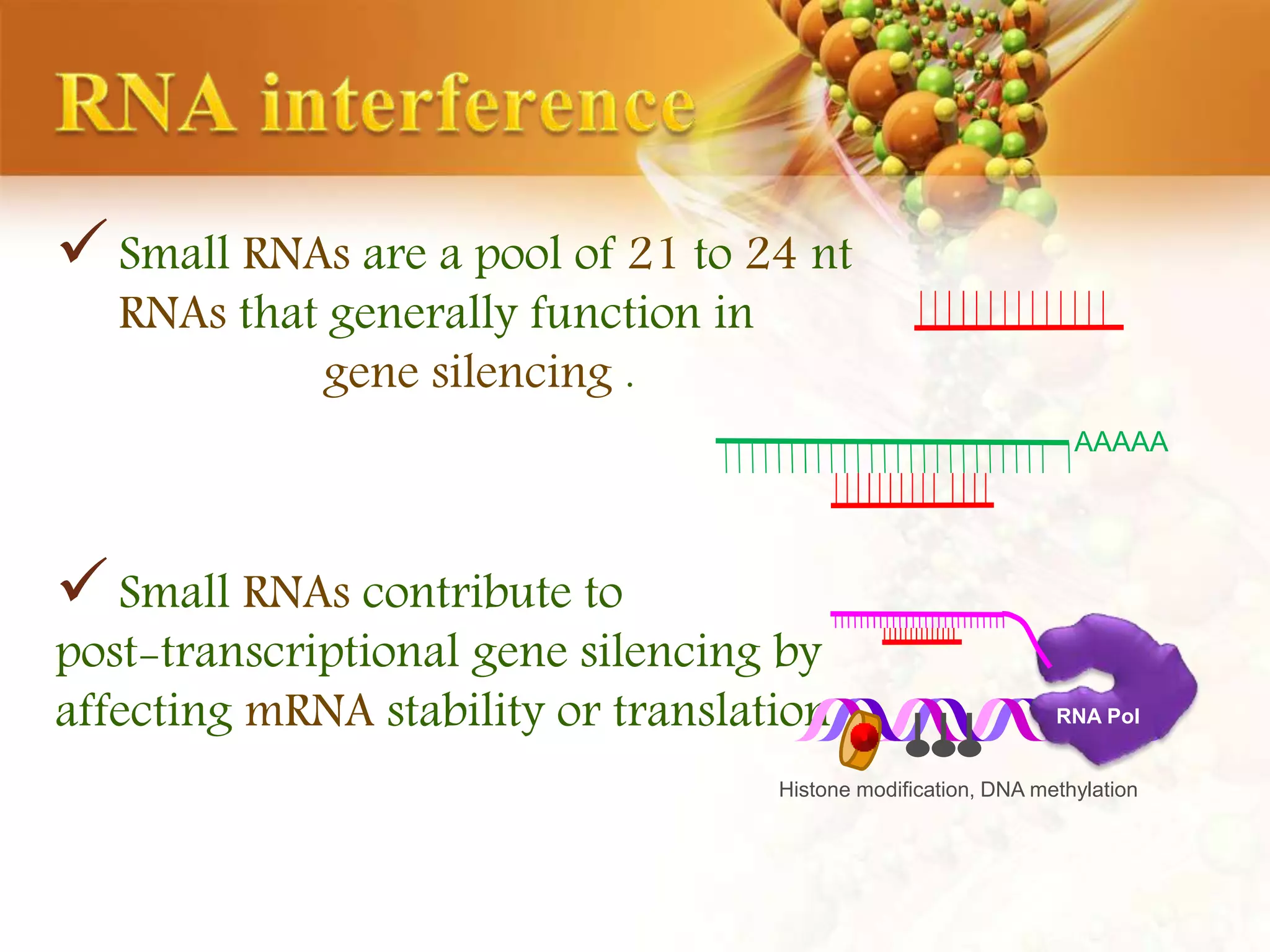
This document discusses genetic engineering techniques used to modify ornamental plants. It describes using Agrobacterium-mediated transformation to insert genes like ipt and barnase-barstar into pelargonium plants. The ipt gene driven by the male specific promoter pSAG12 delayed leaf senescence when expressed. Barnase-barstar induced male sterility. Transgenic plants were selected using antibiotic resistance and validated by PCR and phenotypic analysis. GFP was also used as a visual marker for transformation efficiency. Together these techniques allow genetic engineering of important traits in ornamental crops.

































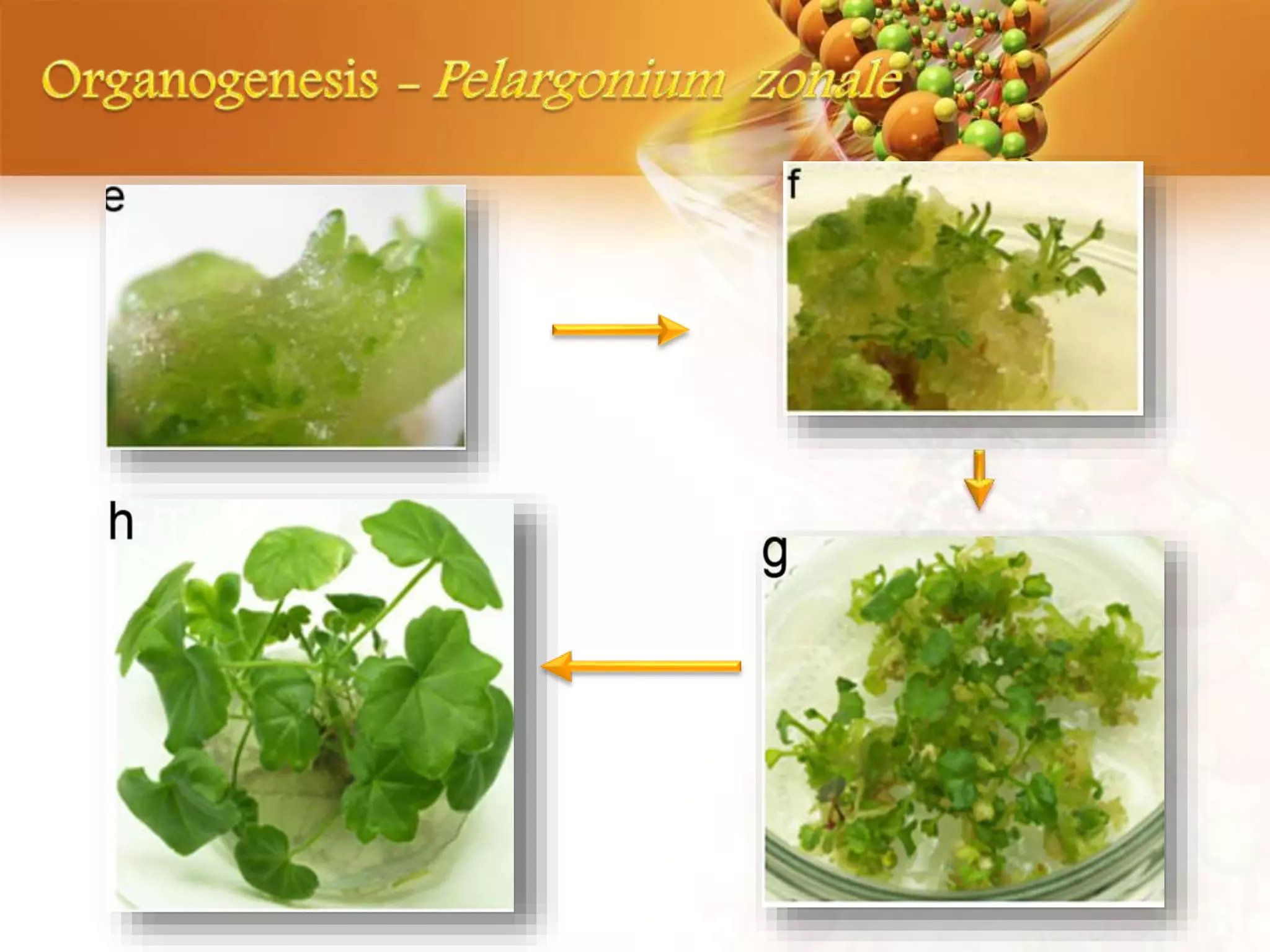
![Plant
material
Surface
sterilization
Calculate
activity cost
drivers’
rates
Rooting
Acclimatization
leaves was cut into 1
cm2 pieces and
cultured on MIM
MS basal medium
and Shahin [46]
vitamins
Morphogen
esis
Induction
Medium
(MIM)
supplemented with
50 mg l-1 kanamycin
Regeneration in
Pelargonium zonale was
carried out via direct
organogenesis and
in Pelargonium peltatum
via somatic
embryogenesis.](https://image.slidesharecdn.com/random-171231165220/75/slide-37-2048.jpg)



![Cytokinins have been implicated in several aspects of
plant development, including plant senescence [15-20],
and are thought to be synthesized mainly in the roots
and transported to the shoots via the xylem.
Overexpression of the ipt gene in transgenic plants led to
elevated foliar cytokinin concentrations and delayed leaf
senescence, but high cytokinin levels have been reported
to be detrimental to growth and fertility [26 30].
To circumvent these effects :
Specificgene promoter (pSAG12 )](https://image.slidesharecdn.com/random-171231165220/75/slide-41-2048.jpg)