RAD51 Drug Discovery (ACS Denver 2011)
•Download as PPT, PDF•
1 like•724 views
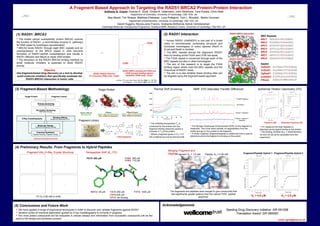
This document describes a fragment-based approach to target the protein-protein interaction between RAD51 and BRCA2 for cancer treatment. Key points: 1) RAD51 and BRCA2 interact through the BRC repeats of BRCA2, which is important for DNA repair. Inhibiting this interaction could block RAD51 activity in tumor cells. 2) The researchers used fragment screening, biophysical techniques like NMR and X-ray crystallography, and chemical elaboration to discover fragments that bind the RAD51-BRCA2 interaction site. 3) Hits were found that bound with low mM affinity. Further optimization through growing and linking fragments yielded compounds with μM affinity, outperforming the natural
Report
Share
Report
Share
Recommended
Fragment Based Drug Discovery

This lecture outlines the different strategies for finding a fragment hit and the subsequent elaboration strategies used in order to increase potency to develop a lead compound in drug discovery.
RecA foscused antibacterial screening

The document discusses the growing problem of antibiotic resistance and potential new approaches to addressing it. It notes that 2 million nosocomial infections occur in the US each year, 70% of which are resistant to at least one drug. 90,000 people die from these infections annually, a nearly 600% increase since 1992. The document then examines RecA, a bacterial protein involved in DNA repair, as a potential new target for antibiotics. It summarizes research investigating various compounds that inhibit RecA's function and could help combat antibiotic resistance.
General Principles of Toxicogenomics

Presentation on Toxicogenomics given by Carole Yauk of the Environmental Health Sciences and Research Bureau of Health Canada in March 2011
MDC Connects: Proteins, structures and how to get them

This document discusses protein production and structure determination. It covers expressing recombinant proteins in common systems like E. coli, insect, and mammalian cells. Purification methods like affinity and size-exclusion chromatography are described. Quality control steps involving SDS-PAGE, mass spectrometry, and functional assays are outlined. Protein crystallography and X-ray crystal structure determination are summarized, including crystallization screening, diffraction data collection, phasing methods, and model building and refinement. The importance of protein structures for drug design is also briefly mentioned.
Ashwini presentation

This document provides an overview of genetic toxicity testing guidelines. It discusses the history and aims of toxicity studies. Various in vitro and in vivo genetic toxicology tests are described, including tests for gene mutation, chromosomal abnormalities, and primary DNA damage. Key tests covered include the mammalian erythrocyte micronucleus test, mammalian bone marrow chromosomal aberration test, rodent dominant lethal assay, and mouse heritable translocation assay. The principles, procedures, and parameters of these tests are summarized. References on genetic toxicology guidance documents and studies are also provided.
MDC Connects: Hit ID screening - understanding your target is key

Domainex is a contract research organization located in Cambridge, UK that provides integrated drug discovery services including medicinal chemistry, protein production and biophysics, and screening. They have experience working with many of the top pharmaceutical companies and have produced over 60 patents and 100 scientific papers. Domainex utilizes a comprehensive approach involving multiple disciplines to identify hits against drug targets. As a case study, they used fragment-based screening and x-ray crystallography to identify small molecule fragments that bind the epigenetic target G9a, which they then expanded into lead compounds.
MDC Connects: Cell-based screening: Old dogs with new tricks

Aurelia Bioscience is a biology CRO that offers various assay development and screening services including:
- NanoBRET assays to study protein-protein interactions and target engagement of kinases
- High throughput screening of over 50,000 compounds using their NanoBRET assays which identified hit compounds
- Target engagement assays using NanoBRET technology to study binding kinetics and residency times of compounds in living cells
- PROTAC drug discovery services using their degradation technology
- High throughput western blotting using the Protein Simple WES system
- Future plans to develop 3D cell-based assays using electrospun scaffold materials.
Biomolecular interaction analysis (BIA) techniques

The document discusses various biomolecular interaction analysis (BIA) techniques for studying interactions between biomolecules and small molecules. It begins with an introduction to BIA and its importance for understanding biology, drug discovery, and diagnostics. The document then outlines and describes several BIA techniques categorized as biochemistry and biophysics methods, molecular methods, computer-aided techniques, and novel creative approaches. Specific techniques discussed in detail include fluorescence spectroscopy, circular dichroism spectroscopy, nuclear magnetic resonance spectroscopy, mass spectrometry, and isothermal titration calorimetry. The document provides information on the principles, applications, advantages, and limitations of each technique.
Recommended
Fragment Based Drug Discovery

This lecture outlines the different strategies for finding a fragment hit and the subsequent elaboration strategies used in order to increase potency to develop a lead compound in drug discovery.
RecA foscused antibacterial screening

The document discusses the growing problem of antibiotic resistance and potential new approaches to addressing it. It notes that 2 million nosocomial infections occur in the US each year, 70% of which are resistant to at least one drug. 90,000 people die from these infections annually, a nearly 600% increase since 1992. The document then examines RecA, a bacterial protein involved in DNA repair, as a potential new target for antibiotics. It summarizes research investigating various compounds that inhibit RecA's function and could help combat antibiotic resistance.
General Principles of Toxicogenomics

Presentation on Toxicogenomics given by Carole Yauk of the Environmental Health Sciences and Research Bureau of Health Canada in March 2011
MDC Connects: Proteins, structures and how to get them

This document discusses protein production and structure determination. It covers expressing recombinant proteins in common systems like E. coli, insect, and mammalian cells. Purification methods like affinity and size-exclusion chromatography are described. Quality control steps involving SDS-PAGE, mass spectrometry, and functional assays are outlined. Protein crystallography and X-ray crystal structure determination are summarized, including crystallization screening, diffraction data collection, phasing methods, and model building and refinement. The importance of protein structures for drug design is also briefly mentioned.
Ashwini presentation

This document provides an overview of genetic toxicity testing guidelines. It discusses the history and aims of toxicity studies. Various in vitro and in vivo genetic toxicology tests are described, including tests for gene mutation, chromosomal abnormalities, and primary DNA damage. Key tests covered include the mammalian erythrocyte micronucleus test, mammalian bone marrow chromosomal aberration test, rodent dominant lethal assay, and mouse heritable translocation assay. The principles, procedures, and parameters of these tests are summarized. References on genetic toxicology guidance documents and studies are also provided.
MDC Connects: Hit ID screening - understanding your target is key

Domainex is a contract research organization located in Cambridge, UK that provides integrated drug discovery services including medicinal chemistry, protein production and biophysics, and screening. They have experience working with many of the top pharmaceutical companies and have produced over 60 patents and 100 scientific papers. Domainex utilizes a comprehensive approach involving multiple disciplines to identify hits against drug targets. As a case study, they used fragment-based screening and x-ray crystallography to identify small molecule fragments that bind the epigenetic target G9a, which they then expanded into lead compounds.
MDC Connects: Cell-based screening: Old dogs with new tricks

Aurelia Bioscience is a biology CRO that offers various assay development and screening services including:
- NanoBRET assays to study protein-protein interactions and target engagement of kinases
- High throughput screening of over 50,000 compounds using their NanoBRET assays which identified hit compounds
- Target engagement assays using NanoBRET technology to study binding kinetics and residency times of compounds in living cells
- PROTAC drug discovery services using their degradation technology
- High throughput western blotting using the Protein Simple WES system
- Future plans to develop 3D cell-based assays using electrospun scaffold materials.
Biomolecular interaction analysis (BIA) techniques

The document discusses various biomolecular interaction analysis (BIA) techniques for studying interactions between biomolecules and small molecules. It begins with an introduction to BIA and its importance for understanding biology, drug discovery, and diagnostics. The document then outlines and describes several BIA techniques categorized as biochemistry and biophysics methods, molecular methods, computer-aided techniques, and novel creative approaches. Specific techniques discussed in detail include fluorescence spectroscopy, circular dichroism spectroscopy, nuclear magnetic resonance spectroscopy, mass spectrometry, and isothermal titration calorimetry. The document provides information on the principles, applications, advantages, and limitations of each technique.
Toxicogenomics review

Toxicogenomics uses gene expression profiling to study how organisms respond to toxic compounds on a global scale. This new approach promises to greatly advance toxicology research. It may help identify toxic mechanisms earlier and assist in predicting compound toxicity. Challenges include interpreting large gene expression datasets and linking changes to specific toxic effects. Progress has been made using toxicogenomics to predict compound mode of action and toxicity pathways. Integrating gene expression data with traditional toxicology can help realize the full potential of this new approach.
Viral and non viral gene transfer

This document discusses gene transfer techniques, including viral and non-viral delivery systems. It describes gene therapy as using genes to treat disease by inserting a gene into a patient's cells instead of using drugs or surgery. It outlines various non-viral physical methods like gene guns, ultrasound, electroporation, and magnetofection and chemical methods like cationic liposomes and polymers to facilitate gene transfer. Viral vectors discussed include retroviruses, adenoviruses, and adeno-associated viruses. In conclusion, while progress has been made, developing safe and effective non-viral delivery systems for in vivo gene therapy remains a challenge.
Stephen Friend Koo Foundation / Sun Yat-Sen Cancer Center 2012-03-12

The document discusses moving beyond linear investigations in science and how we work by integrating layers of omics data models and building computational spaces capable of enabling models to be evolved by teams. It was presented by Stephen Friend of Sage Bionetworks on March 12, 2012 to the Koo Foundation and Sun Yat-Sen Cancer Center.
Applications of Genomic and Proteomic Tools

This document provides an overview of genomic and proteomic tools. It discusses topics like genomics, which is the study of genomes including structural and functional genomics. Proteomics is defined as the large-scale study of proteins, their structures and functions. Several techniques are described briefly, including DNA gel electrophoresis, polymerase chain reaction (PCR), real-time PCR, DNA sequencing, microarray technology, enzyme-linked immunosorbent assay (ELISA), and blotting techniques like Southern blotting, Northern blotting and Western blotting. Applications of these various tools are also mentioned.
SOT short course on computational toxicology 

Computational models are increasingly being used to predict human toxicities. Key enablers of these models include greater availability of data and open source tools. Models have been developed for physicochemical properties, various proteins like cytochromes and transporters, and complex properties like mutagenicity. Future areas of modeling include mitochondrial toxicity, more transporters, nuclear receptors, and green chemistry applications. Wider use of validated open source models and databases is expected, along with mobile applications and more efficient collaboration tools.
MDC Connects: Challenges of Opportunities of Complex Cell Models for Toxicity...

MDC Connects: Challenges of Opportunities of Complex Cell Models for Toxicity...Medicines Discovery Catapult
This document discusses the opportunities and challenges of complex cell models for toxicity testing. It describes how next generation models using stem, primary and CRISPR edited cells in 3D organoids can better mimic the biological environment and improve translation to patients compared to traditional 2D single cell models. Specific complex cell models discussed include a 3D cardiac model using iPSC-derived cardiomyocytes, fibroblasts and endothelial cells to detect cardiotoxicity, as well as blood brain barrier and liver models. While these connected organ-on-chip models show potential, challenges remain around fully defining and validating the models, addressing missing organ systems and targets, achieving scalability and comparability, and demonstrating improved predictivity and translation to human outcomes.MDC Connects Series 2021 | A Guide to Complex Medicines: CryoEM in characteri...

MDC Connects Series 2021 | A Guide to Complex Medicines: CryoEM in characteri...Medicines Discovery Catapult
Our second webinar in the MDC Connects Series 2021 | A Guide to Complex Medicines.
This slide deck takes a closer look at CryoEM in characterisation and quality control of complex medicines
Dr Rebecca Thompson, Astbury Biostructure LaboratoryChEMBL US tour December 2014

This document provides an overview of ChEMBL, a large database of medicinal chemistry data maintained by EMBL-EBI. It describes the types of data contained in ChEMBL, including over 1.6 million compounds, 10,000 targets, and 12 million bioactivities extracted from literature. ChEMBL aims to comprehensively catalogue historical drug discovery successes and failures to identify patterns and support drug discovery. All data is freely available under an open license.
Brian_Strahl 2013_class_on_genomics_and_proteomics

Genomics and Proteomics provides an overview of genomics and proteomics methods and their applications in medicine. It discusses how the fields of genomics and molecular biology emerged through key advances in DNA structure, recombinant DNA technology, PCR, and automated DNA sequencing. The document also reviews epigenetics, genomic and proteomic techniques including blotting, PCR, microarrays, and mass spectrometry. Applications of these methods in clinical settings are described such as genomic tests for disease diagnosis, prognosis, and personalized medicine. Proteomics uses mass spectrometry to discover biomarkers for diseases like cancer.
MDC Connects: Go native… Characterising therapeutic effect in primary cellula...

MDC Connects: Go native… Characterising therapeutic effect in primary cellula...Medicines Discovery Catapult
This document discusses the advantages of using primary cellular models in drug development. Primary cells better mimic native biology and target expression compared to cell lines. Several case studies are presented where primary cellular models helped validate drug targets and identify potential safety issues prior to clinical trials. The document emphasizes characterizing fit-for-purpose models to generate relevant data at each stage of development from discovery to clinical translation. Thorough assay development and validation are important to support clinical applications.Protein protein interaction

Protein-protein interactions are important for many biological processes. There are various types of interactions depending on their composition and duration. Methods to study interactions include yeast two-hybrid, co-immunoprecipitation, affinity chromatography, and chromatin immunoprecipitation. Databases such as IntAct and MINT provide repositories for protein interaction data.
Selection and application of ssDNA aptamers to detect active TB from sputum s...

A paper presentation.
One of the badly-written papers that I've ever read. Seriously.... plenty of speculations and nothing really conclusive.
Metagenomics

Metagenomics is the study of genetic material recovered directly from environmental samples without culturing. This field enables research on uncultured organisms and microbial communities. There are three main metagenomic approaches: biochemical, whole genome shotgun sequencing, and 16s rRNA sequencing. Metagenomics is being applied to study human microbiomes, discover new genes and enzymes, monitor environmental impacts, and characterize uncultured microbes. Future directions include identifying more novel products from uncultured bacteria and improving culture methods and bioinformatics tools.
Site directed mutagenesis

Site-directed mutagenesis is a technique used to introduce specific changes to the DNA sequence of a gene by altering the nucleotide sequence. It allows researchers to study the impact of mutations by changing individual bases, deleting bases, or inserting new bases. There are different methods of site-directed mutagenesis including oligonucleotide-based methods and PCR-based methods. Site-directed mutagenesis has applications in research, production of desired proteins, and development of engineered proteins for commercial uses like detergents.
MICROARRAY

Microarray technology allows researchers to analyze the expression levels of thousands of genes simultaneously using DNA probes attached to a solid surface. There are two main types of microarrays: glass cDNA microarrays which involve spotting pre-fabricated cDNA fragments on glass slides; and high-density oligonucleotide arrays which involve the in situ synthesis of oligonucleotides on a chip. The key steps in a microarray experiment are sample preparation and labeling, hybridization of labeled cDNA to the probes, washing, and image analysis to quantify gene expression levels. Microarrays have numerous applications including gene expression profiling, comparative genomics, disease diagnosis, drug discovery, and toxicology research.
Stephen Friend NIH PPP Coordinating Committee Meeting 2012-02-16

The document discusses using networked team approaches and integrating omics data to build better disease maps through public-private partnerships like CTCAP and Arch2POCM. It proposes sharing clinical and genomic data from comparator arms of trials to create models and de-risking novel drug targets through developing test compounds in a precompetitive space to accelerate new therapies.
Protein-Protein Interactions (PPIs)

The document discusses protein-protein interactions (PPIs), including an introduction to PPIs, the types of interactions, techniques used to study them like X-ray crystallography, NMR spectroscopy and cryo-electron microscopy, and factors that affect PPIs. It also covers methods to investigate PPIs such as affinity purification coupled with mass spectrometry and yeast two-hybrid screening. Applications of understanding PPIs include developing therapeutic drugs and identifying functions of unknown proteins.
SMR kinase meeting October 2013

This document summarizes information about clinical kinase inhibitors and their role in neglected diseases. It discusses establishing a view of the clinical kinome, sources of kinase structure-activity relationship data, community precompetitive efforts like the GSK PKIS dataset, and analysis of kinase inhibitors identified from an HTS screen of compounds active against Mycobacterium tuberculosis. Specific topics covered include the current clinical kinome landscape, top developing companies, attrition rates, polypharmacology profiles, pharmacokinetic data extraction methods, the kinases present in ChEMBL, the GSK PKIS dataset and compound clustering, insights into selectivity/promiscuity, kinases as potential targets in tuberculosis, and docking results suggesting pknB as a target for some
Techniques in proteomics

This document provides an overview of common proteomics techniques. It describes proteomics as the study of proteins including their roles, structures, localization, interactions and other factors. The key techniques discussed include molecular techniques like DNA microarrays and yeast two-hybrid analysis, separation techniques like gel electrophoresis and chromatography, protein identification methods like mass spectroscopy and Edman sequencing, and protein structure determination methods like NMR, X-ray crystallography and computational prediction. The document provides examples and details of several of these techniques.
Proteomics

The document discusses the field of proteomics, which is the large-scale study of proteins, including their functions and structures. It defines proteomics and describes several areas within it, such as functional proteomics, expressional proteomics, and structural proteomics. It outlines typical proteomics experiments and some key methods used, including two-dimensional electrophoresis, mass spectrometry, and protein-protein interaction prediction methods like phylogenetic profiling.
CYP121 Drug Discovery (M. tuberculosis)

Fragment screening was used to target cytochrome P450 (CYP) enzymes from Mycobacterium tuberculosis. Thermal shift assays identified fragment hits against CYP121, which were validated by NMR. X-ray crystallography showed two binding modes. Fragment growing, merging, and linking yielded elaborated fragments with improved binding affinity down to 2 nM against CYP121, maintaining good ligand efficiency. Future work will further optimize compounds against CYP121 and screen other CYP enzymes to develop selective, potent inhibitors to treat tuberculosis.
Graduate lectures (Organic Synthesis in Water)

These two lectures give an overview of organic synthesis using water as a solvent. This is aimed towards final year undergraduates and graduate students in chemistry
More Related Content
What's hot
Toxicogenomics review

Toxicogenomics uses gene expression profiling to study how organisms respond to toxic compounds on a global scale. This new approach promises to greatly advance toxicology research. It may help identify toxic mechanisms earlier and assist in predicting compound toxicity. Challenges include interpreting large gene expression datasets and linking changes to specific toxic effects. Progress has been made using toxicogenomics to predict compound mode of action and toxicity pathways. Integrating gene expression data with traditional toxicology can help realize the full potential of this new approach.
Viral and non viral gene transfer

This document discusses gene transfer techniques, including viral and non-viral delivery systems. It describes gene therapy as using genes to treat disease by inserting a gene into a patient's cells instead of using drugs or surgery. It outlines various non-viral physical methods like gene guns, ultrasound, electroporation, and magnetofection and chemical methods like cationic liposomes and polymers to facilitate gene transfer. Viral vectors discussed include retroviruses, adenoviruses, and adeno-associated viruses. In conclusion, while progress has been made, developing safe and effective non-viral delivery systems for in vivo gene therapy remains a challenge.
Stephen Friend Koo Foundation / Sun Yat-Sen Cancer Center 2012-03-12

The document discusses moving beyond linear investigations in science and how we work by integrating layers of omics data models and building computational spaces capable of enabling models to be evolved by teams. It was presented by Stephen Friend of Sage Bionetworks on March 12, 2012 to the Koo Foundation and Sun Yat-Sen Cancer Center.
Applications of Genomic and Proteomic Tools

This document provides an overview of genomic and proteomic tools. It discusses topics like genomics, which is the study of genomes including structural and functional genomics. Proteomics is defined as the large-scale study of proteins, their structures and functions. Several techniques are described briefly, including DNA gel electrophoresis, polymerase chain reaction (PCR), real-time PCR, DNA sequencing, microarray technology, enzyme-linked immunosorbent assay (ELISA), and blotting techniques like Southern blotting, Northern blotting and Western blotting. Applications of these various tools are also mentioned.
SOT short course on computational toxicology 

Computational models are increasingly being used to predict human toxicities. Key enablers of these models include greater availability of data and open source tools. Models have been developed for physicochemical properties, various proteins like cytochromes and transporters, and complex properties like mutagenicity. Future areas of modeling include mitochondrial toxicity, more transporters, nuclear receptors, and green chemistry applications. Wider use of validated open source models and databases is expected, along with mobile applications and more efficient collaboration tools.
MDC Connects: Challenges of Opportunities of Complex Cell Models for Toxicity...

MDC Connects: Challenges of Opportunities of Complex Cell Models for Toxicity...Medicines Discovery Catapult
This document discusses the opportunities and challenges of complex cell models for toxicity testing. It describes how next generation models using stem, primary and CRISPR edited cells in 3D organoids can better mimic the biological environment and improve translation to patients compared to traditional 2D single cell models. Specific complex cell models discussed include a 3D cardiac model using iPSC-derived cardiomyocytes, fibroblasts and endothelial cells to detect cardiotoxicity, as well as blood brain barrier and liver models. While these connected organ-on-chip models show potential, challenges remain around fully defining and validating the models, addressing missing organ systems and targets, achieving scalability and comparability, and demonstrating improved predictivity and translation to human outcomes.MDC Connects Series 2021 | A Guide to Complex Medicines: CryoEM in characteri...

MDC Connects Series 2021 | A Guide to Complex Medicines: CryoEM in characteri...Medicines Discovery Catapult
Our second webinar in the MDC Connects Series 2021 | A Guide to Complex Medicines.
This slide deck takes a closer look at CryoEM in characterisation and quality control of complex medicines
Dr Rebecca Thompson, Astbury Biostructure LaboratoryChEMBL US tour December 2014

This document provides an overview of ChEMBL, a large database of medicinal chemistry data maintained by EMBL-EBI. It describes the types of data contained in ChEMBL, including over 1.6 million compounds, 10,000 targets, and 12 million bioactivities extracted from literature. ChEMBL aims to comprehensively catalogue historical drug discovery successes and failures to identify patterns and support drug discovery. All data is freely available under an open license.
Brian_Strahl 2013_class_on_genomics_and_proteomics

Genomics and Proteomics provides an overview of genomics and proteomics methods and their applications in medicine. It discusses how the fields of genomics and molecular biology emerged through key advances in DNA structure, recombinant DNA technology, PCR, and automated DNA sequencing. The document also reviews epigenetics, genomic and proteomic techniques including blotting, PCR, microarrays, and mass spectrometry. Applications of these methods in clinical settings are described such as genomic tests for disease diagnosis, prognosis, and personalized medicine. Proteomics uses mass spectrometry to discover biomarkers for diseases like cancer.
MDC Connects: Go native… Characterising therapeutic effect in primary cellula...

MDC Connects: Go native… Characterising therapeutic effect in primary cellula...Medicines Discovery Catapult
This document discusses the advantages of using primary cellular models in drug development. Primary cells better mimic native biology and target expression compared to cell lines. Several case studies are presented where primary cellular models helped validate drug targets and identify potential safety issues prior to clinical trials. The document emphasizes characterizing fit-for-purpose models to generate relevant data at each stage of development from discovery to clinical translation. Thorough assay development and validation are important to support clinical applications.Protein protein interaction

Protein-protein interactions are important for many biological processes. There are various types of interactions depending on their composition and duration. Methods to study interactions include yeast two-hybrid, co-immunoprecipitation, affinity chromatography, and chromatin immunoprecipitation. Databases such as IntAct and MINT provide repositories for protein interaction data.
Selection and application of ssDNA aptamers to detect active TB from sputum s...

A paper presentation.
One of the badly-written papers that I've ever read. Seriously.... plenty of speculations and nothing really conclusive.
Metagenomics

Metagenomics is the study of genetic material recovered directly from environmental samples without culturing. This field enables research on uncultured organisms and microbial communities. There are three main metagenomic approaches: biochemical, whole genome shotgun sequencing, and 16s rRNA sequencing. Metagenomics is being applied to study human microbiomes, discover new genes and enzymes, monitor environmental impacts, and characterize uncultured microbes. Future directions include identifying more novel products from uncultured bacteria and improving culture methods and bioinformatics tools.
Site directed mutagenesis

Site-directed mutagenesis is a technique used to introduce specific changes to the DNA sequence of a gene by altering the nucleotide sequence. It allows researchers to study the impact of mutations by changing individual bases, deleting bases, or inserting new bases. There are different methods of site-directed mutagenesis including oligonucleotide-based methods and PCR-based methods. Site-directed mutagenesis has applications in research, production of desired proteins, and development of engineered proteins for commercial uses like detergents.
MICROARRAY

Microarray technology allows researchers to analyze the expression levels of thousands of genes simultaneously using DNA probes attached to a solid surface. There are two main types of microarrays: glass cDNA microarrays which involve spotting pre-fabricated cDNA fragments on glass slides; and high-density oligonucleotide arrays which involve the in situ synthesis of oligonucleotides on a chip. The key steps in a microarray experiment are sample preparation and labeling, hybridization of labeled cDNA to the probes, washing, and image analysis to quantify gene expression levels. Microarrays have numerous applications including gene expression profiling, comparative genomics, disease diagnosis, drug discovery, and toxicology research.
Stephen Friend NIH PPP Coordinating Committee Meeting 2012-02-16

The document discusses using networked team approaches and integrating omics data to build better disease maps through public-private partnerships like CTCAP and Arch2POCM. It proposes sharing clinical and genomic data from comparator arms of trials to create models and de-risking novel drug targets through developing test compounds in a precompetitive space to accelerate new therapies.
Protein-Protein Interactions (PPIs)

The document discusses protein-protein interactions (PPIs), including an introduction to PPIs, the types of interactions, techniques used to study them like X-ray crystallography, NMR spectroscopy and cryo-electron microscopy, and factors that affect PPIs. It also covers methods to investigate PPIs such as affinity purification coupled with mass spectrometry and yeast two-hybrid screening. Applications of understanding PPIs include developing therapeutic drugs and identifying functions of unknown proteins.
SMR kinase meeting October 2013

This document summarizes information about clinical kinase inhibitors and their role in neglected diseases. It discusses establishing a view of the clinical kinome, sources of kinase structure-activity relationship data, community precompetitive efforts like the GSK PKIS dataset, and analysis of kinase inhibitors identified from an HTS screen of compounds active against Mycobacterium tuberculosis. Specific topics covered include the current clinical kinome landscape, top developing companies, attrition rates, polypharmacology profiles, pharmacokinetic data extraction methods, the kinases present in ChEMBL, the GSK PKIS dataset and compound clustering, insights into selectivity/promiscuity, kinases as potential targets in tuberculosis, and docking results suggesting pknB as a target for some
Techniques in proteomics

This document provides an overview of common proteomics techniques. It describes proteomics as the study of proteins including their roles, structures, localization, interactions and other factors. The key techniques discussed include molecular techniques like DNA microarrays and yeast two-hybrid analysis, separation techniques like gel electrophoresis and chromatography, protein identification methods like mass spectroscopy and Edman sequencing, and protein structure determination methods like NMR, X-ray crystallography and computational prediction. The document provides examples and details of several of these techniques.
Proteomics

The document discusses the field of proteomics, which is the large-scale study of proteins, including their functions and structures. It defines proteomics and describes several areas within it, such as functional proteomics, expressional proteomics, and structural proteomics. It outlines typical proteomics experiments and some key methods used, including two-dimensional electrophoresis, mass spectrometry, and protein-protein interaction prediction methods like phylogenetic profiling.
What's hot (20)
Stephen Friend Koo Foundation / Sun Yat-Sen Cancer Center 2012-03-12

Stephen Friend Koo Foundation / Sun Yat-Sen Cancer Center 2012-03-12
MDC Connects: Challenges of Opportunities of Complex Cell Models for Toxicity...

MDC Connects: Challenges of Opportunities of Complex Cell Models for Toxicity...
MDC Connects Series 2021 | A Guide to Complex Medicines: CryoEM in characteri...

MDC Connects Series 2021 | A Guide to Complex Medicines: CryoEM in characteri...
Brian_Strahl 2013_class_on_genomics_and_proteomics

Brian_Strahl 2013_class_on_genomics_and_proteomics
MDC Connects: Go native… Characterising therapeutic effect in primary cellula...

MDC Connects: Go native… Characterising therapeutic effect in primary cellula...
Selection and application of ssDNA aptamers to detect active TB from sputum s...

Selection and application of ssDNA aptamers to detect active TB from sputum s...
Stephen Friend NIH PPP Coordinating Committee Meeting 2012-02-16

Stephen Friend NIH PPP Coordinating Committee Meeting 2012-02-16
Viewers also liked
CYP121 Drug Discovery (M. tuberculosis)

Fragment screening was used to target cytochrome P450 (CYP) enzymes from Mycobacterium tuberculosis. Thermal shift assays identified fragment hits against CYP121, which were validated by NMR. X-ray crystallography showed two binding modes. Fragment growing, merging, and linking yielded elaborated fragments with improved binding affinity down to 2 nM against CYP121, maintaining good ligand efficiency. Future work will further optimize compounds against CYP121 and screen other CYP enzymes to develop selective, potent inhibitors to treat tuberculosis.
Graduate lectures (Organic Synthesis in Water)

These two lectures give an overview of organic synthesis using water as a solvent. This is aimed towards final year undergraduates and graduate students in chemistry
Prasanth Chikungunya Viral nsP4 

Exploring the Polymerase Activity of Chikungunya Viral non structural Protein 4 (nsP4) using Molecular Modeling, e-Pharmacophore and Docking Studies
S. Prasanth Kumar, Ravi G. Kapopara, Yogesh T. Jasrai and Himanshu A. Pandya
Department of Bioinformatics, ABC, Gujarat University, Ahmedabad- 380009.
In Silico discovery of Metabotropic Glutamate Receptor-3 (mGluR-3) inhibitors

This study developed an in silico drug discovery strategy to identify selective inhibitors of the metabotropic glutamate receptor 3 (mGluR3). The researchers created three pharmacophore models of mGluR3 based on known inhibitors and docking simulations. They screened over 18 million drug-like compounds from the ZINC database using the models, identifying over 130 compounds with predicted binding energies below -9.6 kcal/mol. The top 18 compounds below -10.0 kcal/mol were selected for further analysis in a bioassay to determine potency and selectivity for mGluR3. This in silico approach successfully identified several candidate mGluR3 inhibitor compounds for future experimental testing.
Fragment based screening at inovacia

iNovacia is a drug discovery company that offers fragment-based screening and high-throughput screening services to identify lead compounds. They have a collection of over 280,000 compounds that are screened using NMR, biochemical assays, and other technologies. Hit compounds are then optimized through medicinal chemistry and structural analysis. iNovacia also has expertise in assay development, hit validation, and lead optimization to progress compounds toward drug development.
Fbdd

Fragment-based drug design (FBDD) is an approach to drug discovery that starts with small molecular fragments rather than whole compounds. It identifies fragments that bind to the target protein and then elaborates on those fragments or fuses them together to create lead-like drug molecules. FBDD has advantages over high-throughput screening in that it focuses on developing "lead-like" compounds that are more likely to be optimized into drug candidates. The key steps involve screening a fragment library against the target, elaborating initial fragment hits, and traditional lead optimization methods to generate drug-like molecules.
Fragment_Based_Drug_Design_Abbott

1. A compound was identified that had unique activity against a panel of cancer cell lines but the molecular target was unknown.
2. Analog synthesis defined the structure-activity relationship of the compound.
3. Fragment screening by NMR identified potential binding sites, which was confirmed by X-ray crystallography, and multiple series were progressed to optimization.
FujiFilm Ap 3000

The document advertises the Fujifilm AP-3000, a surface plasmon resonance-based system for label-free fragment-based drug discovery. It can screen 3,840 compounds in 24 hours through fully automated robotics. While previously used, the Fujifilm AP-3000 is now available for sale at surplus pricing by contacting Dragonfly for more information.
NMR in fragment based drug discovery

A presentation discussing fragment-based drug discovery and the development of Abbott drug ABT 263
Webinar: Discovering Small Molecule Protein-Protein Interaction Inhibitors th...

This document advertises a webinar from Computist Bio-Nanotech about designing small molecule protein-protein interaction inhibitors through computational design. The webinar will discuss using computational methods to discover drug compounds that target disease proteins, with examples of designing inhibitors for the Ras/Raf interaction. Computational design could accelerate drug discovery by designing molecules with known modes of action, improving compounds, and making previously difficult targets feasible to drug.
Therapeutic gases

Oxygen is essential for life and deprivation leads most rapidly to death. Oxygen therapy is useful for diseases that interfere with normal oxygenation. Hypoxia refers to insufficient oxygen in tissues and can be caused by problems delivering oxygen to the lungs, abnormal lung function, or inadequate oxygen delivery to tissues. Effects of hypoxia include increased respiration, increased heart rate and blood flow, impaired brain function, and cellular metabolic changes. Oxygen inhalation can reverse hypoxia but excessive amounts can cause toxicity, especially in the central nervous system and lungs.
Protein-protein interaction

The document discusses various strategies for targeting protein-protein interactions (PPIs) for drug development, including small molecule inhibitors, peptides, and allosteric modulation. PPIs are important for many cellular pathways but have been challenging to target with drugs. Examples are given of peptides being developed into peptidomimetics to inhibit PPIs like integrins. Fragment-based approaches and structure-based design have also produced PPI inhibitors for targets like interleukin-2 receptor. Covalent modifiers and stabilizing allosteric sites are additional strategies discussed.
Molecular Dynamics

The document provides an overview of molecular dynamics (MD) simulations. It discusses the history of MD, including early contributions from Alder and Wainwright in the 1950s-1980s studying interactions of hard spheres. The document outlines the MD algorithm, which uses Newton's second law and numerical integration methods like the Euler method to calculate particle trajectories based on interaction potentials over time. It also discusses some common uses and limitations of MD simulations, such as their ability to bridge microscopic and macroscopic scales but potential for unexpected results due to approximations.
Ecosystems PowerPoint Presentation

The document summarizes and compares two ecosystems in Puerto Rico: Bosque Seco de Guánica and El Yunque National Forest. It describes the different forest areas within each ecosystem, including the types of trees and canopy layers. It also discusses the differences in rainfall, soil composition, and how each ecosystem has adapted. Finally, it provides details on some of the unique flora and fauna found in each forest and some environmental problems currently facing them.
Viewers also liked (14)
In Silico discovery of Metabotropic Glutamate Receptor-3 (mGluR-3) inhibitors

In Silico discovery of Metabotropic Glutamate Receptor-3 (mGluR-3) inhibitors
Webinar: Discovering Small Molecule Protein-Protein Interaction Inhibitors th...

Webinar: Discovering Small Molecule Protein-Protein Interaction Inhibitors th...
Similar to RAD51 Drug Discovery (ACS Denver 2011)
Pjb Probes 2009

The document outlines the goals of the Epigenetics Project to discover chemical probes for epigenetic targets. Significant progress has been made, including developing probes for the HMT G9a and bromodomain-containing proteins in the BET subfamily. Assays have been established for many epigenetic protein families with several probes in development. Collaborations with academic and pharmaceutical partners have contributed screening capabilities and medicinal chemistry support.
MSc Final Project - Alvaro Diaz Mendoza

The document summarizes a study that investigated the interaction between the Hfq protein from E. coli bacteria and DNA. Specifically, it examined the binding of two regions of Hfq - the carboxyl terminal region (CTR) and mutations in the amino terminal region (NTR) - to DNA using techniques like isothermal titration calorimetry and electrophoretic mobility shift assays. The results showed that both the CTR and mutated NTR proteins bound to DNA to some degree. Atomic force microscopy was also used to characterize the self-assembly behavior of CTR peptides in order to aid interpretation of the calorimetry data.
Medicilon KRAS-targeted Drugs R&D Service.pdf

RAS is one of the most frequently mutated oncogenes in human cancer. KRAS is the isoform most frequently mutated, which constitutes about 85% of RAS mutations. As the most frequently mutated RAS isoform, KRAS is intensively studied in the past years.
In the formulation of KRAS integrated research plan, Medicilon has in-depth communication with customers. The backbone of scientific research has combined the characteristics of each case with years of practical experience and technical accumulation, and carefully submitted high-quality experimental plans and results to customers. Medicilon provides KRAS-targeted drug discovery, CMC research (API + formulation), pharmacodynamics research, PK study, safety evaluation and other services.https://www.medicilon.com/platform/kras/
Gene Expression Analysis by Real Time PCR

Nowadays, Real Time-PCR analysis has become the method of choice for gene expression studies and validating transcriptomic data.
Isolation of promoter and other regulatory elements.pptx

I apologize, I do not have enough context to answer questions about molecular biology concepts or experiments. My role is to summarize written documents.
Microarray validation

This document describes how real-time PCR can be used to validate microarray data. Real-time PCR provides a quantitative and sensitive method for confirming changes in gene expression observed in microarray experiments. The document outlines a protocol for designing and running a real-time PCR experiment to validate a specific result from a microarray experiment showing increased expression of the TNFAIP3 gene in response to TNFα treatment. Key steps in the protocol include performing reverse transcription of RNA to generate cDNA, setting up a standard curve and controls, and analyzing the real-time PCR data to calculate fold-changes in gene expression.
Aacr poster2007

This document summarizes a study that used pathway-focused real-time PCR arrays to study extracellular matrix proteins and cell adhesion molecules in breast tumor development. Two PCR arrays were used - a Cancer PathwayFinder array and an Extracellular Matrix and Adhesion Molecules array. RNA from a breast tumor and normal breast tissue were analyzed on the Cancer PathwayFinder array, identifying 33 genes with at least a 3-fold expression change between tumor and normal tissue, including 7 cell adhesion molecules. Further analysis of a second tumor on the Extracellular Matrix array identified 38 cell adhesion genes with at least a 3-fold change, implicating these pathways in breast tumor formation.
New-gene manipulication class -xs 2023-课件1(周一).pdf

This document discusses gene manipulation and genomic analysis techniques. It begins with an introduction to the speaker's educational and employment background working in biology. The document then outlines the topics to be covered in the course, including recombinant DNA principles and applications, as well as different gene manipulation and genomic analysis techniques that students will discuss in groups. Finally, it provides an overview of various DNA cloning, screening, and sequencing techniques used in genetic engineering and genomic research.
defense 2.0

This document summarizes research on engineering bivalent affinity ligands. Key points include:
- A combinatorial bivalent ligand library was generated that identified ligands binding non-overlapping epitopes and maintained thermostability. This library performed better than a monovalent library.
- The best ligands from the bivalent library were sequenced, with 9 found to be bivalent and 1 trivalent. Mutations outside the original library positions were found in some ligands.
- Ligand M2 was identified that bound with low nanomolar affinity and could engage in multi-epitope binding to intrinsically disordered domains of beta-catenin. M2 showed promise as a new binding scaffold.
- Dimer
Thesis Intro for LinkedIn

This document introduces the topic of using a molecular beacon probe called KRAS G12V to identify a single-nucleotide polymorphism (SNP) in the oncogene KRAS. It will explore developing an assay using linear-after-the-exponential PCR (LATE-PCR) and the molecular beacon probe to distinguish the KRAS G12V SNP from other KRAS genotypes. It provides background information on real-time quantitative PCR, LATE-PCR, and molecular beacon probes to set up exploring the development of this new assay.
IRJET- Silencing of hnRNP A1 and hnRNP A2/B1 Downregulates the Expression of ...

The document summarizes a study that examined the effect of silencing hnRNP A1 and hnRNP A2/B1 splice factors on the expression of CD44v6 and CD44v10 exons in glioma cells. The researchers found that knockdown of hnRNP A1 and hnRNP A2/B1 led to decreased expression of CD44v6 and CD44v10 exons based on qRT-PCR analysis. Specifically, cells with silenced splice factors had lower expression of the two exons compared to non-silenced control cells. Therefore, hnRNP A1 and hnRNP A2/B1 may positively regulate the expression of
Structure Prediction of WDR13 and a study of its Interacting Partners

Structure Prediction of WDR13 using Homology Modelling and Fold Recognition and a study of it's interacting partners using automated docking.
Basler modellers.210126reduced

I presented two fascinating stories where Molecular Dynamics simulations contributed to enhancing our understanding of immunodeficiencies. In one of the projects, the treatment of patients could be improved. These slides were presented at the Basler Modeller Stammtisch, 26.02.2021
The Assembly, Structure and Activation of Influenza a M2 Transmembrane Domain...

This document summarizes two research papers on computational methods for analyzing protein structures and interactions. The first paper describes a Bayesian method for determining protein structures from sparse single-molecule X-ray diffraction data. The second paper presents xMDFF, a new molecular dynamics flexible fitting approach for refining low-resolution protein structures determined by X-ray crystallography. The third paper introduces i-ATTRACT, a new flexible protein-protein docking method that combines rigid body and flexible interface residue energy minimization for predicting protein complex structures.
ASCB_Poster_edit_TM1_GA1_TV5

- The researchers created homology models of the Mitotic Checkpoint Complex (MCC) proteins Cdc20p, Mad2p, and Mad3p from budding yeast using an existing crystal structure from fission yeast as a template.
- Molecular dynamics simulations suggested the cdc20-1 mutation, which causes a temperature sensitive phenotype, does not significantly impact the protein's thermodynamic stability.
- The data indicates the mutation likely causes defects in protein folding kinetics rather than directly impacting protein binding domains or stability.
Cook2010web

The document provides an overview of computational chemistry methods for structure-activity relationship analysis, pharmacophore modeling, and protein-ligand docking. It discusses topics like SAR, QSAR, molecular alignment, conformational analysis, homology modeling of protein targets, and docking programs. Examples are given of applying these methods to study benzodiazepine ligands and GABA receptor subtypes.
Sequencing the Human TCRβ Repertoire on the Ion S5TM

understanding of the human immune system, and thereby cancer immunology.
αβT-cells are the primary constituents of human cell-mediated adaptive immunity.
The antigen specificity of each αβT-cell is encoded in the 500-600 bp transcript
encompassing the variable portion of the rearranged TCRα and TCRβ subunits,
which can be read via NGS in a process termed repertoire sequencing. Until now,
the main challenge the field faces is the lack of a technology that can provide a
contiguous read of 600 bp to minimize the complexity of designing bias-prone
primers and informatics challenges of stitching short reads. Here we leverage the
long read capability of Ion 530™ chip to comprehensively sequence all three CDR
domains of the TCRβ chain. The Ion 530™ chip offers greater than 15 M productive
reads, allowing a multiplex of 2-4 samples with sufficient coverage for most repertoire
profiling studies. Initial testing with leukocyte total RNA demonstrates that this
multiplex PCR assay produced repertoires that were much more similar to data
derived from 5’-RACE protocol than the commonly used BIOMED-2 primer set. This
result suggested that the use of long reads minimizes bias by allowing targeting of
less variable regions. To further assess the performance of the assay, we designed a
model system of 30 plasmid controls containing common human T-cell CDR3
sequences. Each plasmid was amplified individually and sequenced to confirm the
detection of a single clonal population. Analytical sensitivity of the assay and
accuracy of the accompanied analysis solution were further evaluated by spiking in
plasmid concentrations from 10 pg to 0.0001 pg (5 million to 50 copies) in a
background of 100 ng cDNA reverse transcribed from leukocyte total RNA. Results
showed the assay offers linearity over 5 orders of magnitude of decreasing input
concentration. In summary, we have demonstrated a NGS workflow for TCRβ
sequencing that offers multiplex flexibility on Ion S5 with sample to answer in less
than 48 hours.
The Butterfly Effect: How to see the impact of small changes to your ADC

This document summarizes key aspects of characterizing antibody drug conjugates (ADCs), including:
1) A case study examining how different polyethylene glycol (PEG) linker sizes affect ADC structure and target binding. Peptide mapping by mass spectrometry showed conjugation sites varied with linker size. Hydrogen/deuterium exchange mass spectrometry showed conjugation induced conformational changes.
2) Methods for assessing ADC mechanisms of action, including measuring internalization, cytotoxicity, and effector functions.
3) An overview of MilliporeSigma's comprehensive ADC product characterization and biosafety testing services across multiple sites.
The Butterfly Effect: How to see the impact of small changes to your ADC

Watch this webinar here: https://bit.ly/31PRr2z
Small changes to the design of antibody-drug conjugate can have a dramatic effect on its structure and biological activity. Effective product characterization is essential to understanding the impact of these changes. Here we discuss methods to provide insight at critical junctures in ADC development.
There are many different design considerations facing developers of antibody-drug conjugates: these variables must be tuned to achieve the right balance of efficacy and safety. For example, the choice of linker can influence an ADC's potency, toxicity and pharmacokinetics.
In this webinar we explore the influence of various PEG linkers on the structure of a model ADC by identifying specific sites of conjugation by peptide mapping, investigating changes in higher order structure by HDX mass spectrometry, and examining the impact on binding by SPR spectroscopy.
We demonstrate that employing a range of orthogonal methods is critical to understanding the structure-function relationships of an ADC.
In this webinar, you will learn about:
• How the choice of linker can influence an ADC's activity
• Information-rich methods to probe ADC structure and function
• Effective strategies for thorough characterization of ADC products
Q biomarkersomaticmutation

The document summarizes qBiomarker Somatic Mutation PCR Arrays, which are PCR-based assays that can rapidly and accurately detect somatic mutations in cancer samples. The arrays can detect mutations present at as little as 0.01% of DNA in a sample. They have been validated to work reliably with different sample types, including archived samples. The arrays cover a wide range of clinically relevant cancer mutations and allow screening of many mutations simultaneously in a single PCR run. The assays and content were selected based on published data on mutation frequencies and functional significance. The arrays provide a simple method for sensitive somatic mutation profiling to aid cancer research.
Similar to RAD51 Drug Discovery (ACS Denver 2011) (20)
Isolation of promoter and other regulatory elements.pptx

Isolation of promoter and other regulatory elements.pptx
New-gene manipulication class -xs 2023-课件1(周一).pdf

New-gene manipulication class -xs 2023-课件1(周一).pdf
IRJET- Silencing of hnRNP A1 and hnRNP A2/B1 Downregulates the Expression of ...

IRJET- Silencing of hnRNP A1 and hnRNP A2/B1 Downregulates the Expression of ...
Structure Prediction of WDR13 and a study of its Interacting Partners

Structure Prediction of WDR13 and a study of its Interacting Partners
The Assembly, Structure and Activation of Influenza a M2 Transmembrane Domain...

The Assembly, Structure and Activation of Influenza a M2 Transmembrane Domain...
Sequencing the Human TCRβ Repertoire on the Ion S5TM

Sequencing the Human TCRβ Repertoire on the Ion S5TM
The Butterfly Effect: How to see the impact of small changes to your ADC

The Butterfly Effect: How to see the impact of small changes to your ADC
The Butterfly Effect: How to see the impact of small changes to your ADC

The Butterfly Effect: How to see the impact of small changes to your ADC
Recently uploaded
Bob Reedy - Nitrate in Texas Groundwater.pdf

Presented at June 6-7 Texas Alliance of Groundwater Districts Business Meeting
Deep Behavioral Phenotyping in Systems Neuroscience for Functional Atlasing a...

Functional Magnetic Resonance Imaging (fMRI) provides means to characterize brain activations in response to behavior. However, cognitive neuroscience has been limited to group-level effects referring to the performance of specific tasks. To obtain the functional profile of elementary cognitive mechanisms, the combination of brain responses to many tasks is required. Yet, to date, both structural atlases and parcellation-based activations do not fully account for cognitive function and still present several limitations. Further, they do not adapt overall to individual characteristics. In this talk, I will give an account of deep-behavioral phenotyping strategies, namely data-driven methods in large task-fMRI datasets, to optimize functional brain-data collection and improve inference of effects-of-interest related to mental processes. Key to this approach is the employment of fast multi-functional paradigms rich on features that can be well parametrized and, consequently, facilitate the creation of psycho-physiological constructs to be modelled with imaging data. Particular emphasis will be given to music stimuli when studying high-order cognitive mechanisms, due to their ecological nature and quality to enable complex behavior compounded by discrete entities. I will also discuss how deep-behavioral phenotyping and individualized models applied to neuroimaging data can better account for the subject-specific organization of domain-general cognitive systems in the human brain. Finally, the accumulation of functional brain signatures brings the possibility to clarify relationships among tasks and create a univocal link between brain systems and mental functions through: (1) the development of ontologies proposing an organization of cognitive processes; and (2) brain-network taxonomies describing functional specialization. To this end, tools to improve commensurability in cognitive science are necessary, such as public repositories, ontology-based platforms and automated meta-analysis tools. I will thus discuss some brain-atlasing resources currently under development, and their applicability in cognitive as well as clinical neuroscience.
Nucleic Acid-its structural and functional complexity.

This presentation explores a brief idea about the structural and functional attributes of nucleotides, the structure and function of genetic materials along with the impact of UV rays and pH upon them.
The use of Nauplii and metanauplii artemia in aquaculture (brine shrimp).pptx

Although Artemia has been known to man for centuries, its use as a food for the culture of larval organisms apparently began only in the 1930s, when several investigators found that it made an excellent food for newly hatched fish larvae (Litvinenko et al., 2023). As aquaculture developed in the 1960s and ‘70s, the use of Artemia also became more widespread, due both to its convenience and to its nutritional value for larval organisms (Arenas-Pardo et al., 2024). The fact that Artemia dormant cysts can be stored for long periods in cans, and then used as an off-the-shelf food requiring only 24 h of incubation makes them the most convenient, least labor-intensive, live food available for aquaculture (Sorgeloos & Roubach, 2021). The nutritional value of Artemia, especially for marine organisms, is not constant, but varies both geographically and temporally. During the last decade, however, both the causes of Artemia nutritional variability and methods to improve poorquality Artemia have been identified (Loufi et al., 2024).
Brine shrimp (Artemia spp.) are used in marine aquaculture worldwide. Annually, more than 2,000 metric tons of dry cysts are used for cultivation of fish, crustacean, and shellfish larva. Brine shrimp are important to aquaculture because newly hatched brine shrimp nauplii (larvae) provide a food source for many fish fry (Mozanzadeh et al., 2021). Culture and harvesting of brine shrimp eggs represents another aspect of the aquaculture industry. Nauplii and metanauplii of Artemia, commonly known as brine shrimp, play a crucial role in aquaculture due to their nutritional value and suitability as live feed for many aquatic species, particularly in larval stages (Sorgeloos & Roubach, 2021).
Cytokines and their role in immune regulation.pptx

This presentation covers the content and information on "Cytokines " and their role in immune regulation .
The debris of the ‘last major merger’ is dynamically young

The Milky Way’s (MW) inner stellar halo contains an [Fe/H]-rich component with highly eccentric orbits, often referred to as the
‘last major merger.’ Hypotheses for the origin of this component include Gaia-Sausage/Enceladus (GSE), where the progenitor
collided with the MW proto-disc 8–11 Gyr ago, and the Virgo Radial Merger (VRM), where the progenitor collided with the
MW disc within the last 3 Gyr. These two scenarios make different predictions about observable structure in local phase space,
because the morphology of debris depends on how long it has had to phase mix. The recently identified phase-space folds in Gaia
DR3 have positive caustic velocities, making them fundamentally different than the phase-mixed chevrons found in simulations
at late times. Roughly 20 per cent of the stars in the prograde local stellar halo are associated with the observed caustics. Based
on a simple phase-mixing model, the observed number of caustics are consistent with a merger that occurred 1–2 Gyr ago.
We also compare the observed phase-space distribution to FIRE-2 Latte simulations of GSE-like mergers, using a quantitative
measurement of phase mixing (2D causticality). The observed local phase-space distribution best matches the simulated data
1–2 Gyr after collision, and certainly not later than 3 Gyr. This is further evidence that the progenitor of the ‘last major merger’
did not collide with the MW proto-disc at early times, as is thought for the GSE, but instead collided with the MW disc within
the last few Gyr, consistent with the body of work surrounding the VRM.
Thornton ESPP slides UK WW Network 4_6_24.pdf

ESPP presentation to EU Waste Water Network, 4th June 2024 “EU policies driving nutrient removal and recycling
and the revised UWWTD (Urban Waste Water Treatment Directive)”
SAR of Medicinal Chemistry 1st by dk.pdf

In this presentation include the prototype drug SAR on thus or with their examples .
Syllabus of Second Year B. Pharmacy
2019 PATTERN.
Unlocking the mysteries of reproduction: Exploring fecundity and gonadosomati...

The pygmy halfbeak Dermogenys colletei, is known for its viviparous nature, this presents an intriguing case of relatively low fecundity, raising questions about potential compensatory reproductive strategies employed by this species. Our study delves into the examination of fecundity and the Gonadosomatic Index (GSI) in the Pygmy Halfbeak, D. colletei (Meisner, 2001), an intriguing viviparous fish indigenous to Sarawak, Borneo. We hypothesize that the Pygmy halfbeak, D. colletei, may exhibit unique reproductive adaptations to offset its low fecundity, thus enhancing its survival and fitness. To address this, we conducted a comprehensive study utilizing 28 mature female specimens of D. colletei, carefully measuring fecundity and GSI to shed light on the reproductive adaptations of this species. Our findings reveal that D. colletei indeed exhibits low fecundity, with a mean of 16.76 ± 2.01, and a mean GSI of 12.83 ± 1.27, providing crucial insights into the reproductive mechanisms at play in this species. These results underscore the existence of unique reproductive strategies in D. colletei, enabling its adaptation and persistence in Borneo's diverse aquatic ecosystems, and call for further ecological research to elucidate these mechanisms. This study lends to a better understanding of viviparous fish in Borneo and contributes to the broader field of aquatic ecology, enhancing our knowledge of species adaptations to unique ecological challenges.
3D Hybrid PIC simulation of the plasma expansion (ISSS-14)

3D Particle-In-Cell (PIC) algorithm,
Plasma expansion in the dipole magnetic field.
20240520 Planning a Circuit Simulator in JavaScript.pptx

Evaporation step counter work. I have done a physical experiment.
(Work in progress.)
Sharlene Leurig - Enabling Onsite Water Use with Net Zero Water

Sharlene Leurig - Enabling Onsite Water Use with Net Zero WaterTexas Alliance of Groundwater Districts
Presented at June 6-7 Texas Alliance of Groundwater Districts Business MeetingApplied Science: Thermodynamics, Laws & Methodology.pdf

When I was asked to give a companion lecture in support of ‘The Philosophy of Science’ (https://shorturl.at/4pUXz) I decided not to walk through the detail of the many methodologies in order of use. Instead, I chose to employ a long standing, and ongoing, scientific development as an exemplar. And so, I chose the ever evolving story of Thermodynamics as a scientific investigation at its best.
Conducted over a period of >200 years, Thermodynamics R&D, and application, benefitted from the highest levels of professionalism, collaboration, and technical thoroughness. New layers of application, methodology, and practice were made possible by the progressive advance of technology. In turn, this has seen measurement and modelling accuracy continually improved at a micro and macro level.
Perhaps most importantly, Thermodynamics rapidly became a primary tool in the advance of applied science/engineering/technology, spanning micro-tech, to aerospace and cosmology. I can think of no better a story to illustrate the breadth of scientific methodologies and applications at their best.
Shallowest Oil Discovery of Turkiye.pptx

The Petroleum System of the Çukurova Field - the Shallowest Oil Discovery of Türkiye, Adana
The binding of cosmological structures by massless topological defects

Assuming spherical symmetry and weak field, it is shown that if one solves the Poisson equation or the Einstein field
equations sourced by a topological defect, i.e. a singularity of a very specific form, the result is a localized gravitational
field capable of driving flat rotation (i.e. Keplerian circular orbits at a constant speed for all radii) of test masses on a thin
spherical shell without any underlying mass. Moreover, a large-scale structure which exploits this solution by assembling
concentrically a number of such topological defects can establish a flat stellar or galactic rotation curve, and can also deflect
light in the same manner as an equipotential (isothermal) sphere. Thus, the need for dark matter or modified gravity theory is
mitigated, at least in part.
BREEDING METHODS FOR DISEASE RESISTANCE.pptx

Plant breeding for disease resistance is a strategy to reduce crop losses caused by disease. Plants have an innate immune system that allows them to recognize pathogens and provide resistance. However, breeding for long-lasting resistance often involves combining multiple resistance genes
Deep Software Variability and Frictionless Reproducibility

Deep Software Variability and Frictionless ReproducibilityUniversity of Rennes, INSA Rennes, Inria/IRISA, CNRS
The ability to recreate computational results with minimal effort and actionable metrics provides a solid foundation for scientific research and software development. When people can replicate an analysis at the touch of a button using open-source software, open data, and methods to assess and compare proposals, it significantly eases verification of results, engagement with a diverse range of contributors, and progress. However, we have yet to fully achieve this; there are still many sociotechnical frictions.
Inspired by David Donoho's vision, this talk aims to revisit the three crucial pillars of frictionless reproducibility (data sharing, code sharing, and competitive challenges) with the perspective of deep software variability.
Our observation is that multiple layers — hardware, operating systems, third-party libraries, software versions, input data, compile-time options, and parameters — are subject to variability that exacerbates frictions but is also essential for achieving robust, generalizable results and fostering innovation. I will first review the literature, providing evidence of how the complex variability interactions across these layers affect qualitative and quantitative software properties, thereby complicating the reproduction and replication of scientific studies in various fields.
I will then present some software engineering and AI techniques that can support the strategic exploration of variability spaces. These include the use of abstractions and models (e.g., feature models), sampling strategies (e.g., uniform, random), cost-effective measurements (e.g., incremental build of software configurations), and dimensionality reduction methods (e.g., transfer learning, feature selection, software debloating).
I will finally argue that deep variability is both the problem and solution of frictionless reproducibility, calling the software science community to develop new methods and tools to manage variability and foster reproducibility in software systems.
Exposé invité Journées Nationales du GDR GPL 2024
Recently uploaded (20)
Deep Behavioral Phenotyping in Systems Neuroscience for Functional Atlasing a...

Deep Behavioral Phenotyping in Systems Neuroscience for Functional Atlasing a...
Nucleic Acid-its structural and functional complexity.

Nucleic Acid-its structural and functional complexity.
The use of Nauplii and metanauplii artemia in aquaculture (brine shrimp).pptx

The use of Nauplii and metanauplii artemia in aquaculture (brine shrimp).pptx
Cytokines and their role in immune regulation.pptx

Cytokines and their role in immune regulation.pptx
The debris of the ‘last major merger’ is dynamically young

The debris of the ‘last major merger’ is dynamically young
Unlocking the mysteries of reproduction: Exploring fecundity and gonadosomati...

Unlocking the mysteries of reproduction: Exploring fecundity and gonadosomati...
3D Hybrid PIC simulation of the plasma expansion (ISSS-14)

3D Hybrid PIC simulation of the plasma expansion (ISSS-14)
aziz sancar nobel prize winner: from mardin to nobel

aziz sancar nobel prize winner: from mardin to nobel
20240520 Planning a Circuit Simulator in JavaScript.pptx

20240520 Planning a Circuit Simulator in JavaScript.pptx
Sharlene Leurig - Enabling Onsite Water Use with Net Zero Water

Sharlene Leurig - Enabling Onsite Water Use with Net Zero Water
Applied Science: Thermodynamics, Laws & Methodology.pdf

Applied Science: Thermodynamics, Laws & Methodology.pdf
The binding of cosmological structures by massless topological defects

The binding of cosmological structures by massless topological defects
Deep Software Variability and Frictionless Reproducibility

Deep Software Variability and Frictionless Reproducibility
RAD51 Drug Discovery (ACS Denver 2011)
- 1. A Fragment Based Approach to Targeting the RAD51:BRCA2 Protein-Protein Interaction Anthony G. Coyne, Duncan E. Scott, Chiara R. Valenzano, John Skidmore, Tara Pukala, Chris Abell Department of Chemistry, University of Cambridge, CB2 1EW, UK May Marsh, Tim Sharpe, Matthias Ehebauer, Luca Pellegrini, Tom L. Blundell , Marko Hyvonen Department of Biochemistry, University of Cambridge, CB2 1GA, UK. David Huggins, Nicola-Jane Francis, Grahame McKenzie Ashok Venkitaraman Cambridge Molecular Therapeutics Programme, Hutchison/MRC Research Centre, University of Cambridge, CB2 0XZ, UK. (1) RAD51: BRCA2 (2) RAD51 Interaction (4) Preliminary Results: From Fragments to Hybrid Peptides (3) Fragment-Based Methodology (5) Conclusions and Future Work Acknowledgements QuickTime™ and a decompressor are needed to see this picture. QuickTime™ and a decompressor are needed to see this picture. Seeding Drug Discovery Initiative: GR 091058 Translation Award: GR 080083 The breast cancer susceptibility protein BRCA2 controls the function of RAD51, a recombinase enzyme in pathways for DNA repair by homologus recombination. BRCA2 binds RAD51 through eight BRC repeats and an overexpression of the BRC4 repeat in cells prevents formation of RAD51-ssDNA nucleofilament and results in RAD51 diffusion and failure to repair DNA breaks The disruption on the RAD51-BRCA2 binding interface by small molecule inhibitors is expected to block RAD51 activity. Objective Use fragment-based drug discovery as a tool to develop small molecule inhibitors that specifically modulate the RAD51-BRCA2 interaction in tumor cells Human RAD51 (HsRAD51) is one part of a broad class of recombinases comprising structural and functional homologues in every species (RecA in E.coli and RadA in Archea) The BRC repeats disrupt the oligomeric RAD51 form by binding at the FXXA and LFDE hot spots The FXXA repeat is conserved through each of the BRC repeats but also in other homologues The aim of this research is to target the FXXA binding region where both the BRC repeats and the N-terminal of RAD51 binds The aim is to see whether these binding sites can be targeted using the fragment-based approach RAD51-BRC4 Interaction RAD51-RAD51 Interaction BRC Repeats BRC1 HSFGGSFRTASNKEI BRC2 EVGFRGFYSAHGTKL BRC3 ETSDTFFQTASGKNI BRC4 EPTLLGFHTASGKKV BRC6 EVGPPAFRIASGKIV BRC7 ANTCGIFSTASGKSV BRC8 SSAFSGFSTASGKQV RAD51 RAD51-RAD51 Filament (S. Cerevisiae (PDB Code: 1szp))a QuickTime™ and a decompressor are needed to see this picture. RAD51-BRC4 showing the FXXA and LFDE hot-spot binding regions (HsRAD51 (PDB Code: 1now)b LFDE FXXA QuickTime™ and a decompressor are needed to see this picture. (a) Rice, P.A. et al, Nat. Struct. Mol. Biol, 2004, 11, 791-796 (b) Pellegrini L.. et al, Nature, 2002, 420, 287-293 Target Protein Fragment Library Primary Screening Thermal Shift Screening Secondary Screening NMR Spectroscopy X-Ray Crystallography Binding Affinity Isothermal Calorimetry (ITC) Molecular Design Fragment Analoguing, Docking Chemical Synthesis Fragment Growing, Fragment Linking Iterative Development Cycle Target Protein Thermal Shift Screening NMR: STD (Saturated Transfer Difference) Isothermal Titration Calorimetry (ITC) RAD51-BRCA2 Binding site PfRadA Green: Identical Residues Red: Different Residues PfRadA Mutant Green: Identical Residues Red: Different Residues Fragment Library OH N O N N N O HO CF3 N O O O HN O O O O OH O H N NH2N O HO N S 1338 Fragments 0 5000 10000 15000 20000 25 30 35 40 45 50 55 Temperature (o C) The unfolding temperature TM is monitored for the protein and any fragment binding observed causes a increase in TM of the protein Where a fragment gives a ∆TM >1o C this is defined as a hit by thermal shift The Nuclear Overhauser Enhancement (NOE) on the fragment is measured. This is the direct transfer of magnetization from the methyl groups on the protein to the fragment A known displacer (fragment/peptide) is added and this is used to determine where the fragment is binding on the protein 1 H NMR Fragment No Protein Protein+ Fragment Protein+ Fragment + Displacer ITC measures the heat released or absorbed during ligand binding to the protein. The binding constant (KB), n (stoichiometry) ∆H and ∆S can all be calculated from this experiment. Fragment (mM) Elaborated Fragment (nM) ITC KD 0.56 mM to 2mM email: agc40@cam.ac.uk QuickTime™ and a decompressor are needed to see this picture. QuickTime™ and a decompressor are needed to see this picture. QuickTime™ and a decompressor are needed to see this picture. QuickTime™ and a decompressor are needed to see this picture. Tetrapeptide SAR (KD ITC)Fragment Hits (X-Ray Crystal Structure) Merging Fragment and Peptide Phe Pocket Ala Pocket Fragment: KD = 1.4 mM Peptide: KD = 0.28 mM We have applied a range of biophysical techniques in order to discover and validate fragments against RAD51 Iterative cycles of chemical elaboration guided by X-ray crystallography is currently in progress The most potent compounds will be evaluated in cellular assays and information from successful compounds will be fed back to the design and synthesis process KD = 5.9 µM KD = 2.6 µM Fragment/Peptide Hybrid 1 Fragment/Peptide Hybrid 2 FATA 280 µM FNTA 630 µM FPTA No binding WHTA 95 µM FHTG 1000 µM FHPA 110 µM FHAA 500 µMFHTA 280 µM The fragments and peptides were merged to give compounds that had significantly greater potency than the natural FXXA peptide sequence KLVPMGFTTATEFHQ RLVPMGFVTAADFHM KLVPMGFTTATEFHQ KLVPLGFLSARTFYQ AANLGTFMRADEYLK HsRad51 ScRad51 GgRad51 DmRad51 PfRadA
