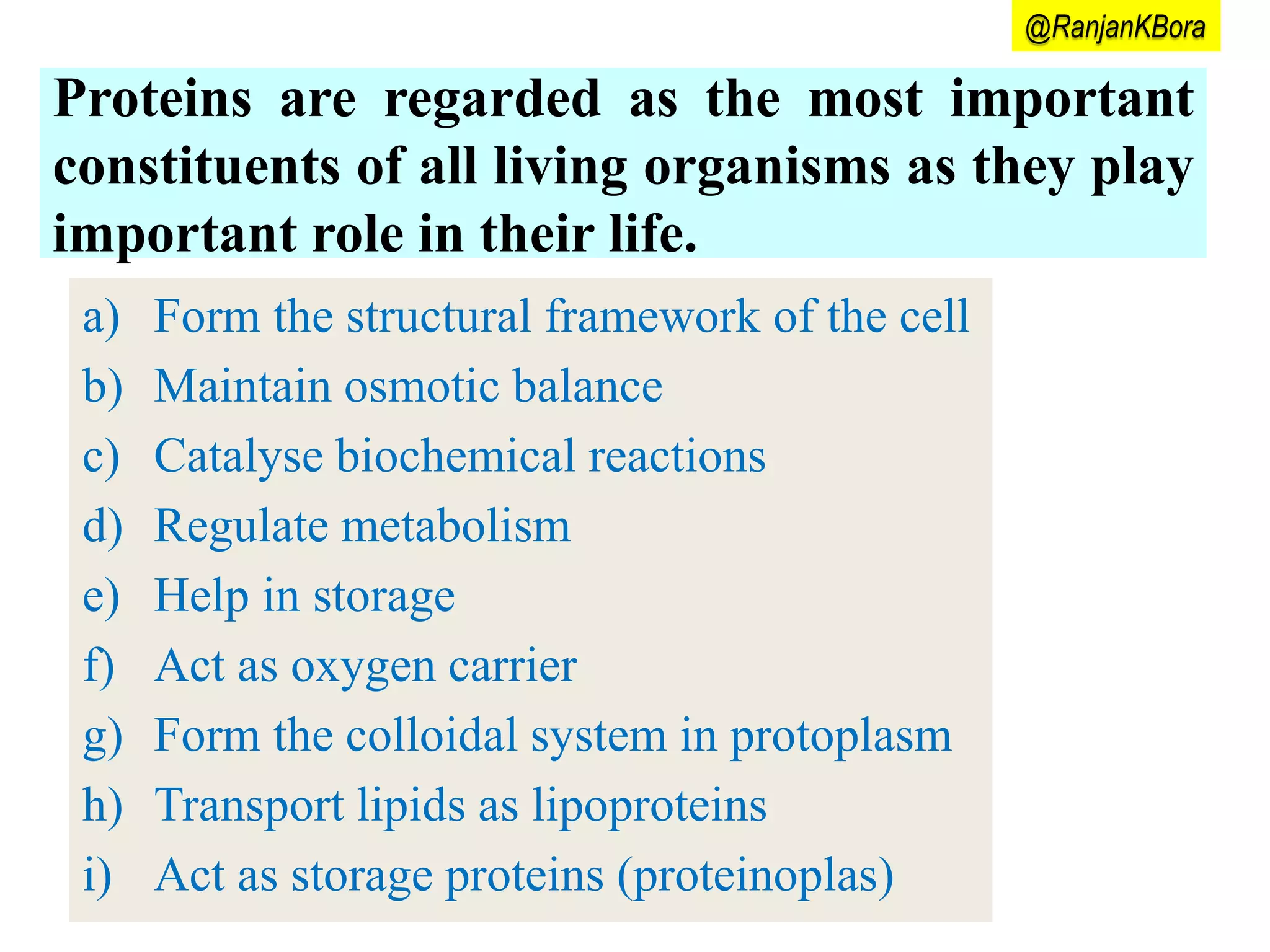
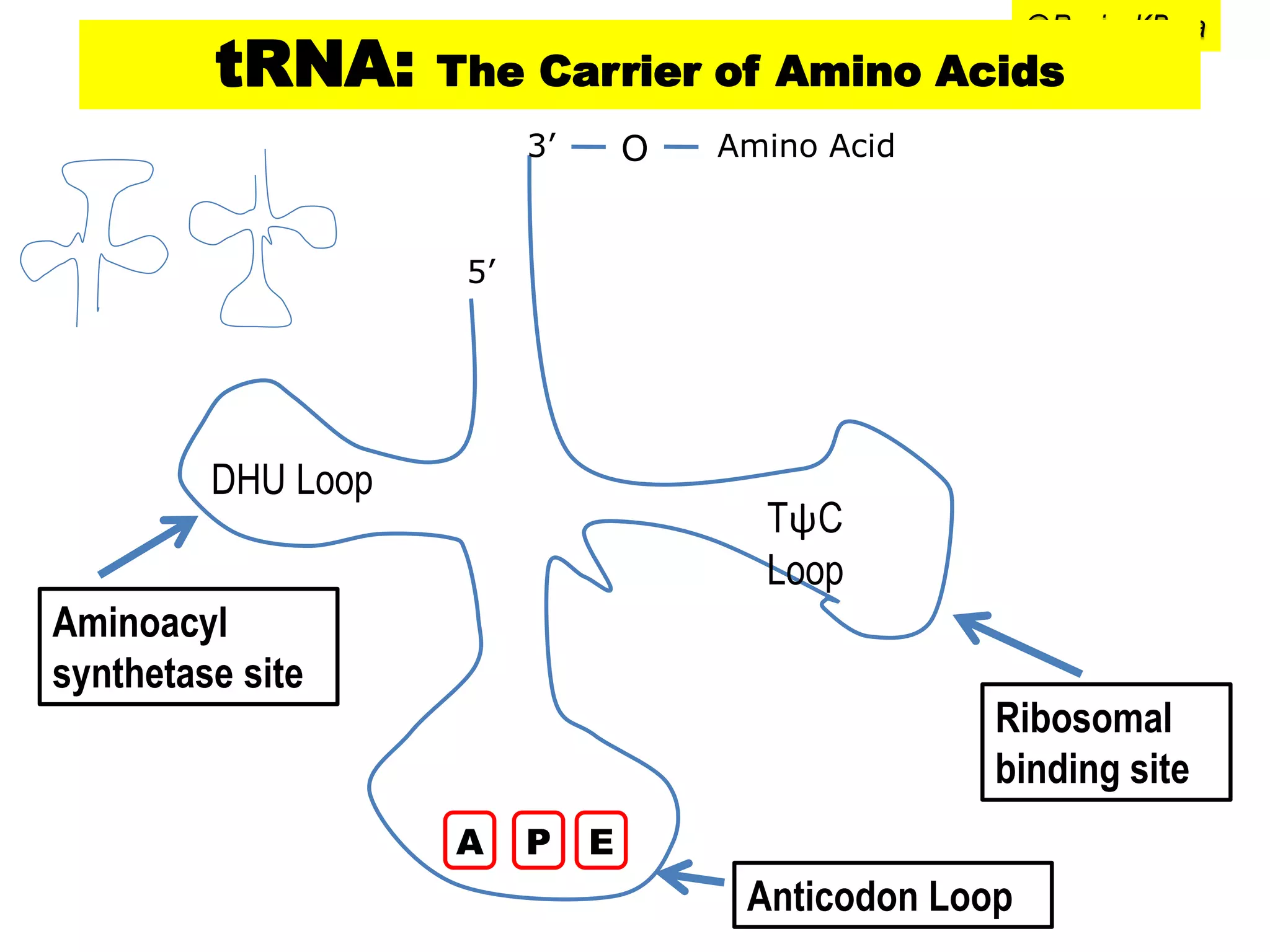
Proteins play important roles in living organisms as structural components and catalysts. The central dogma of molecular biology describes the flow of genetic information from DNA to RNA to protein. Transcription and translation are the two processes by which the information in DNA is used to synthesize proteins. Transcription involves copying DNA into mRNA which is then translated into protein with the help of tRNA and the ribosome. Translation begins with initiation and involves elongation and termination steps to assemble the amino acid sequence specified by the mRNA.