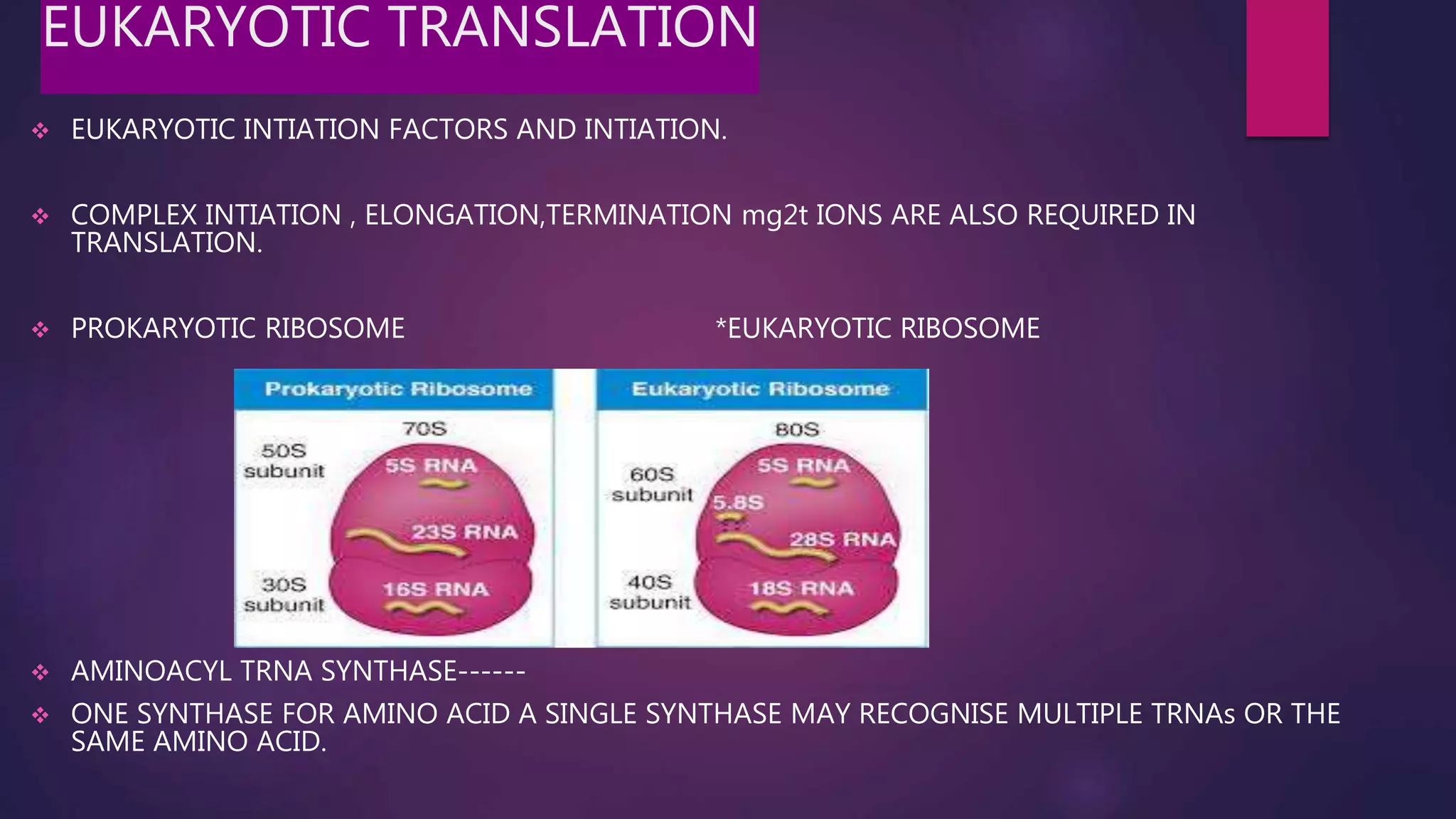
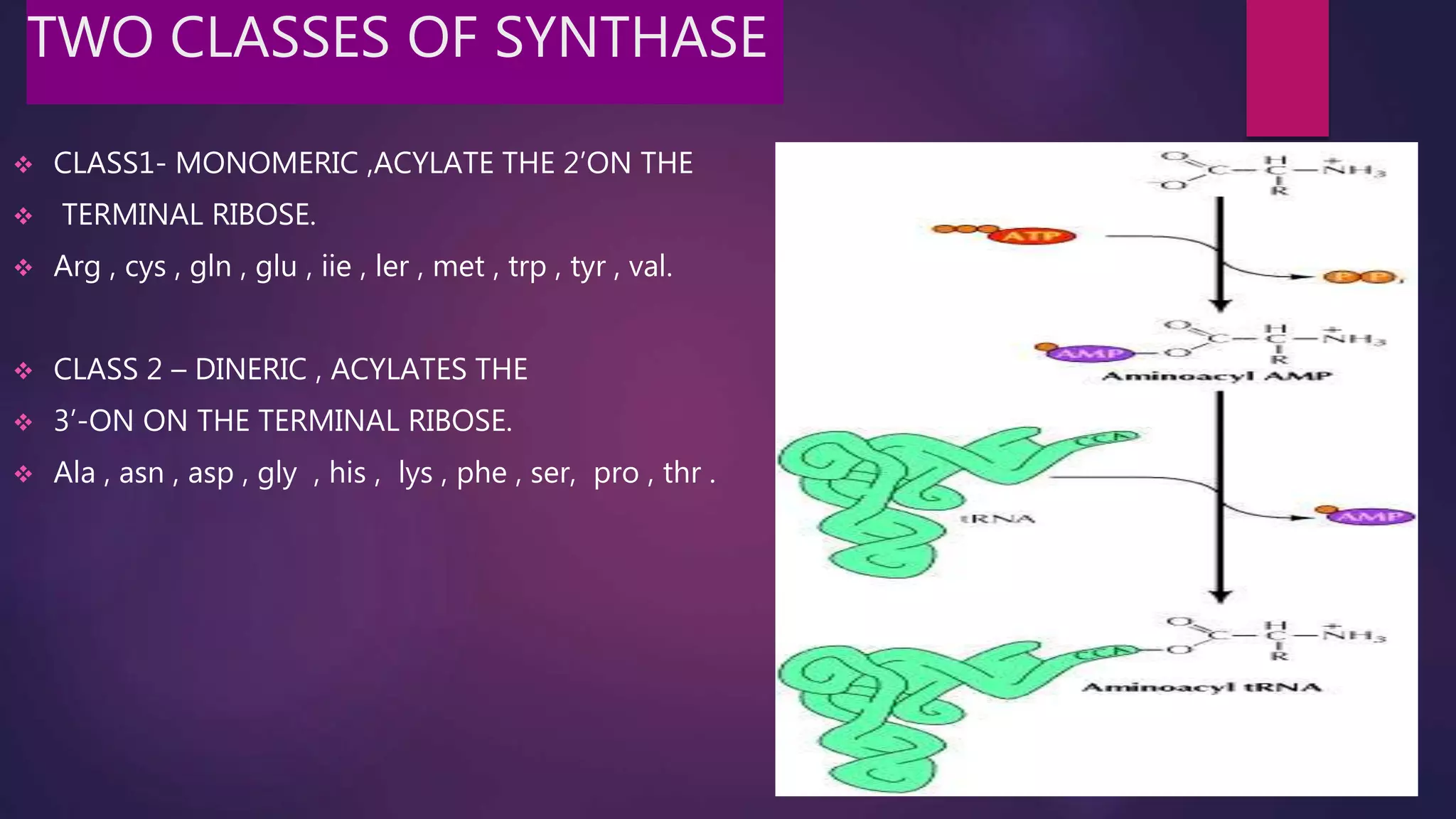
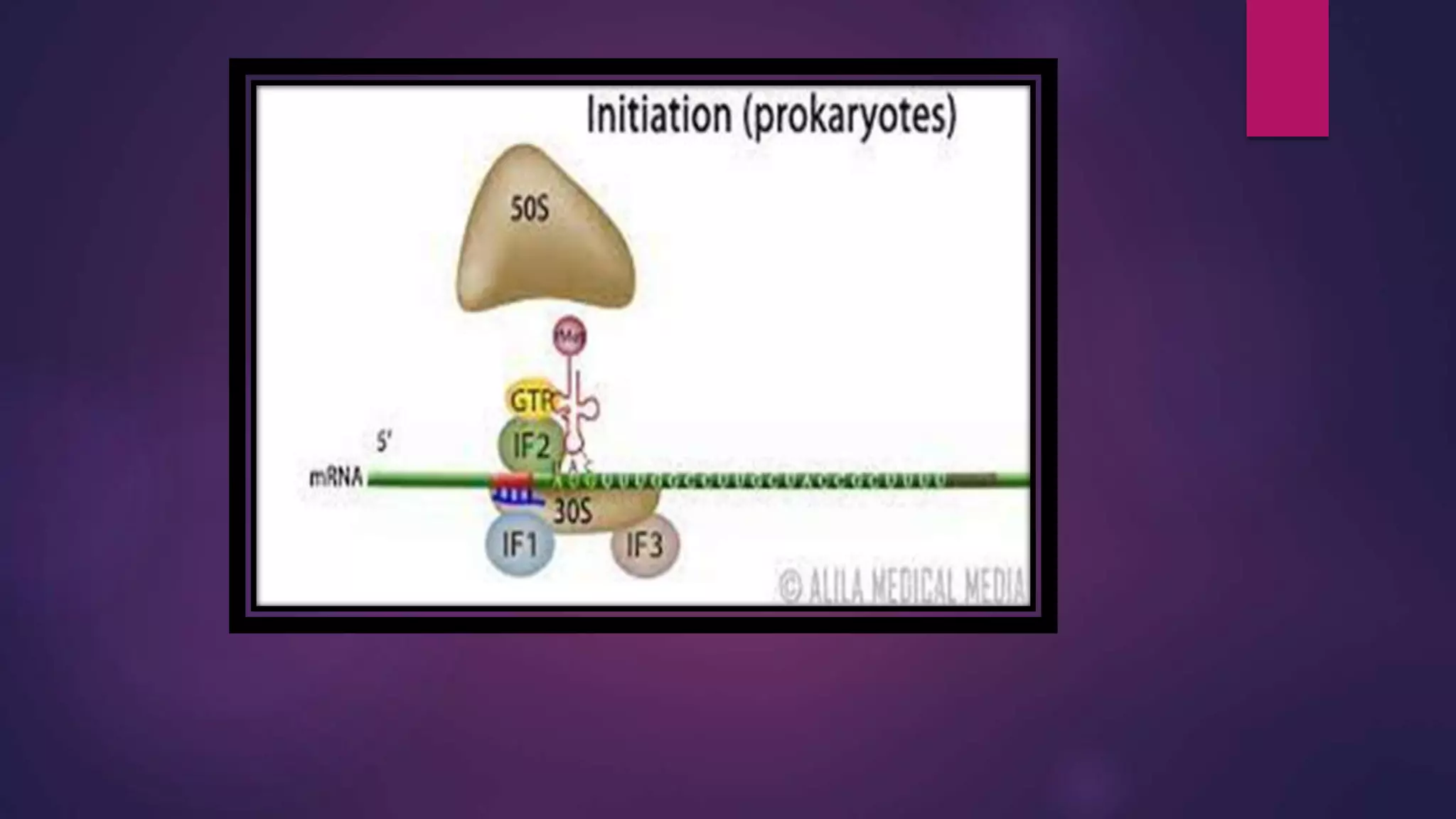
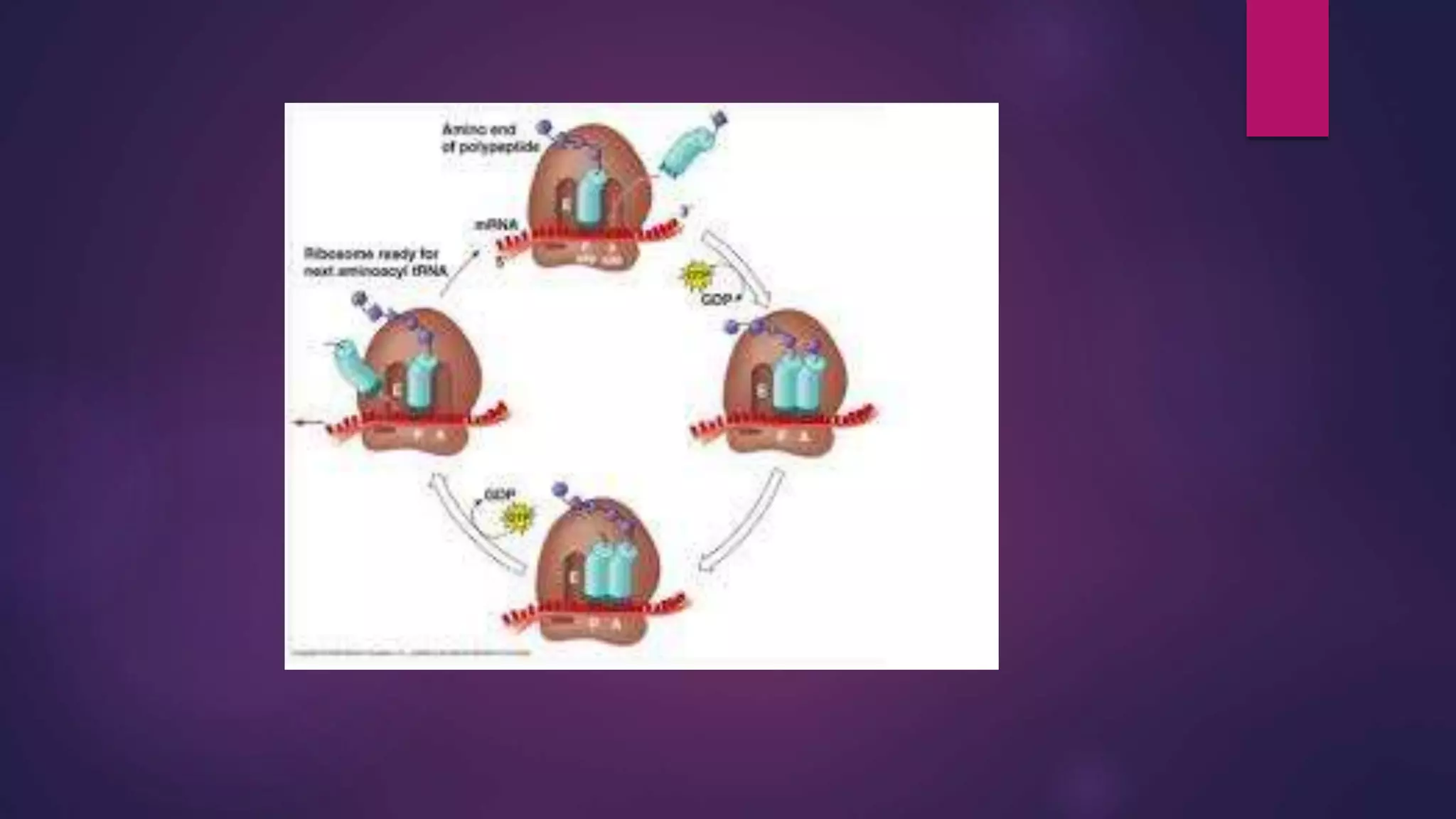
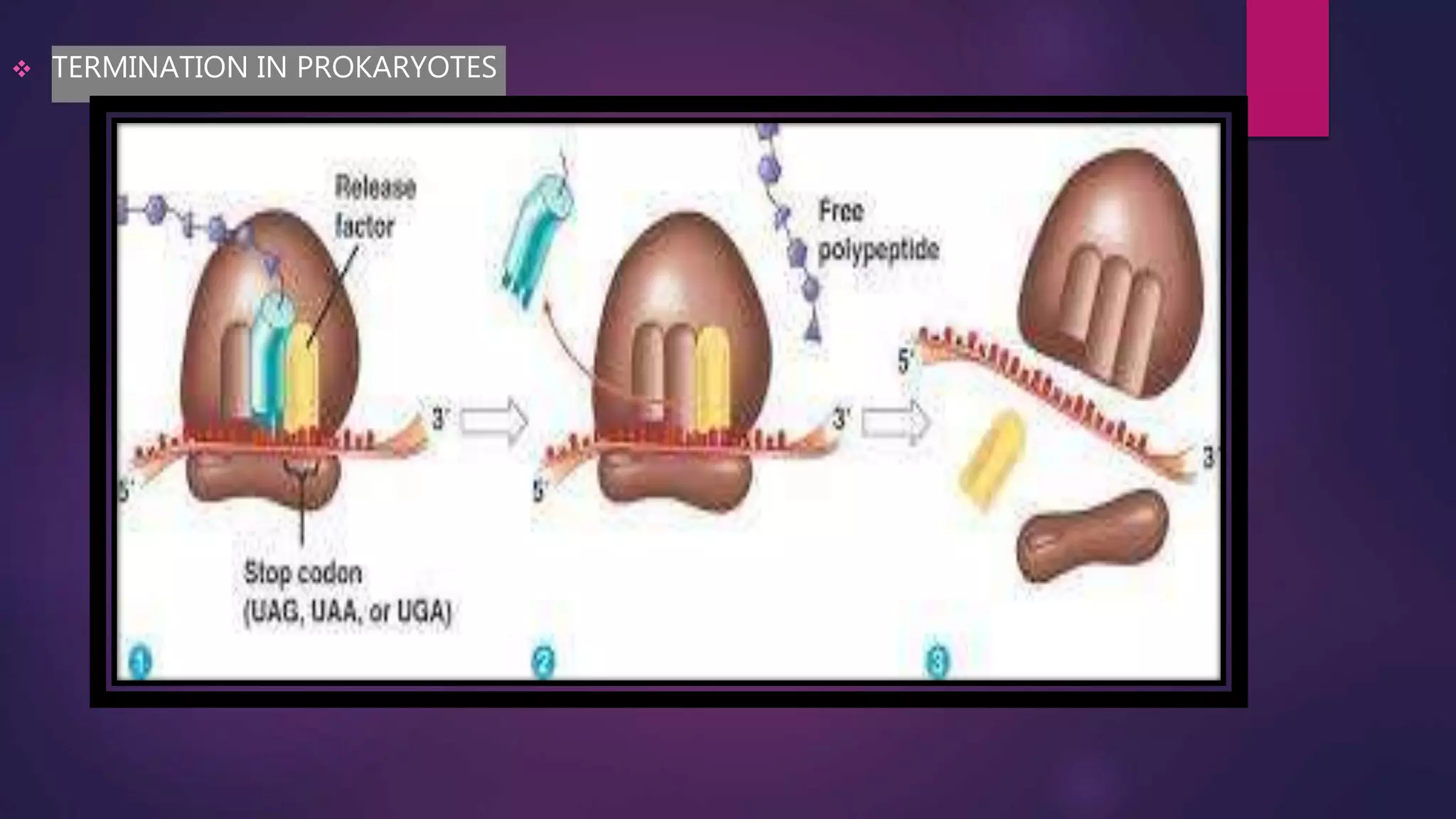
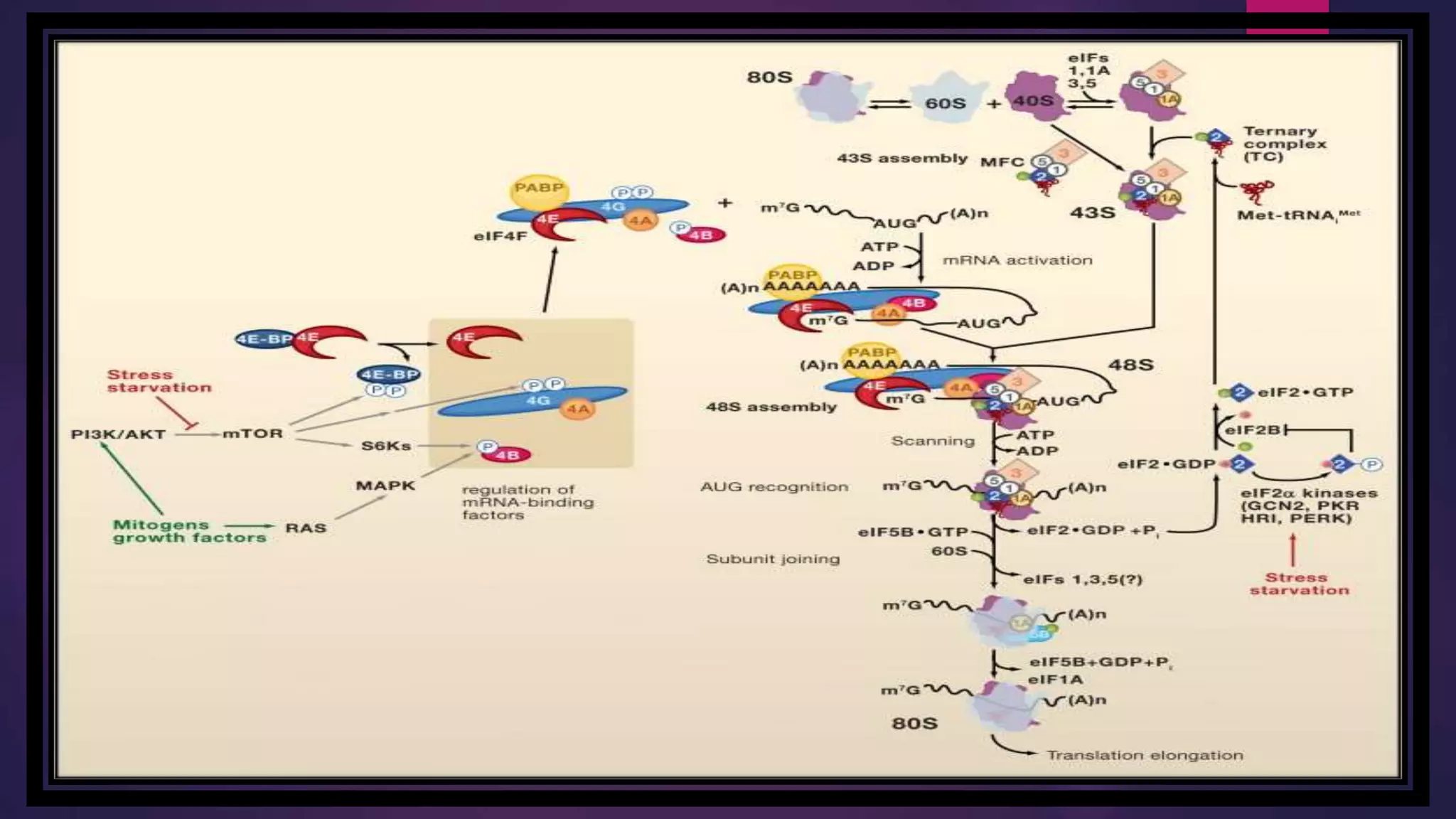
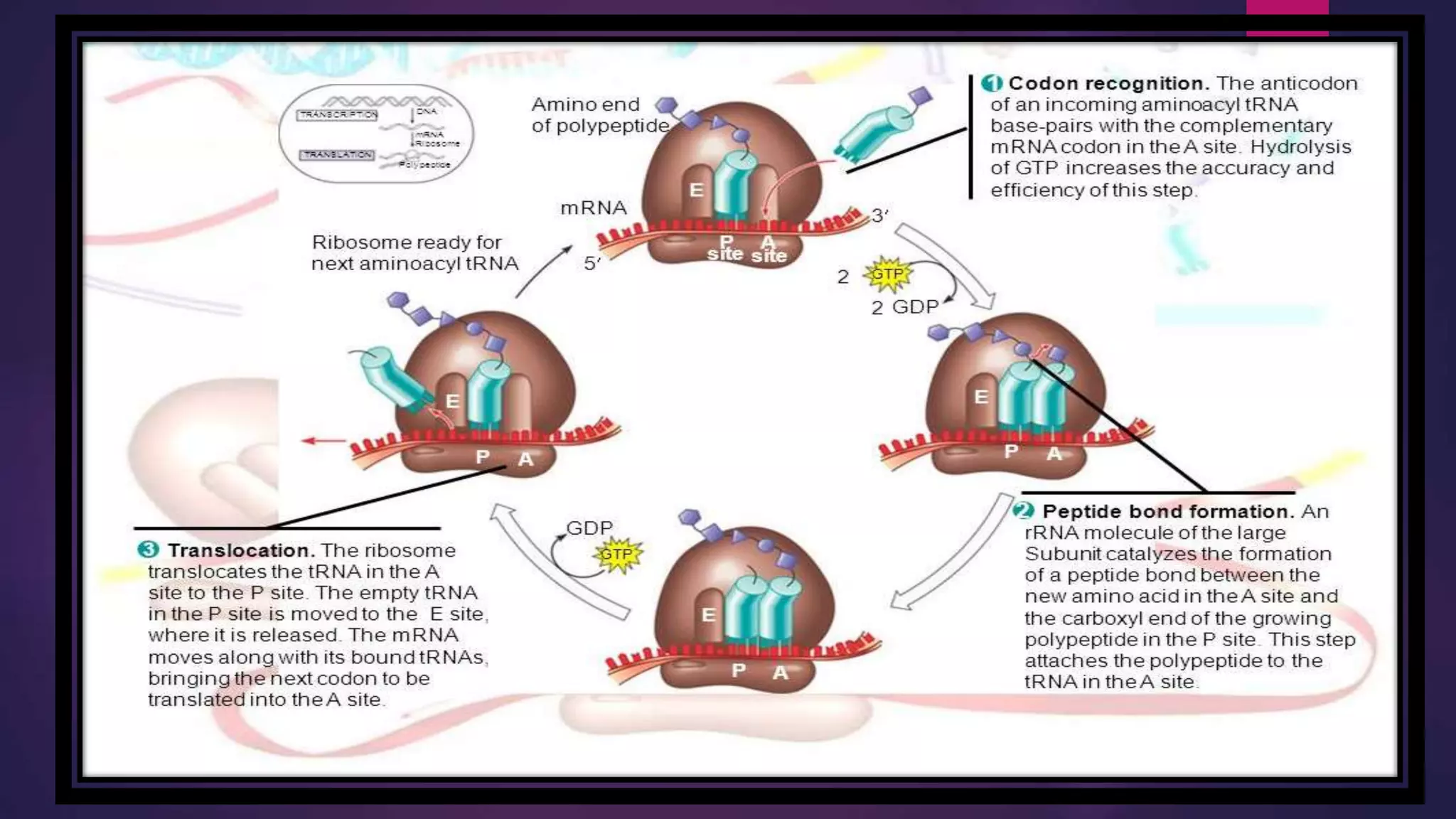
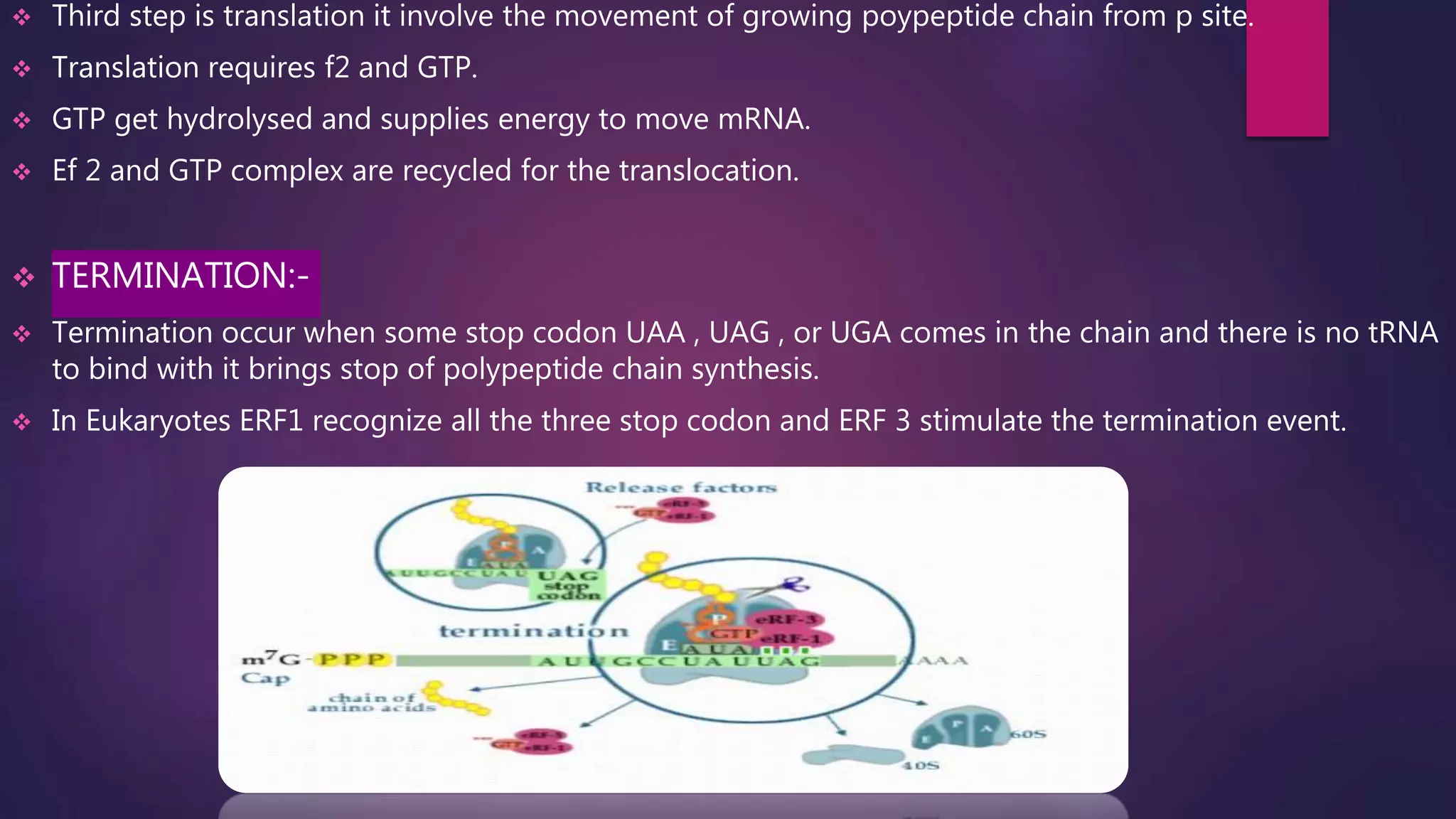
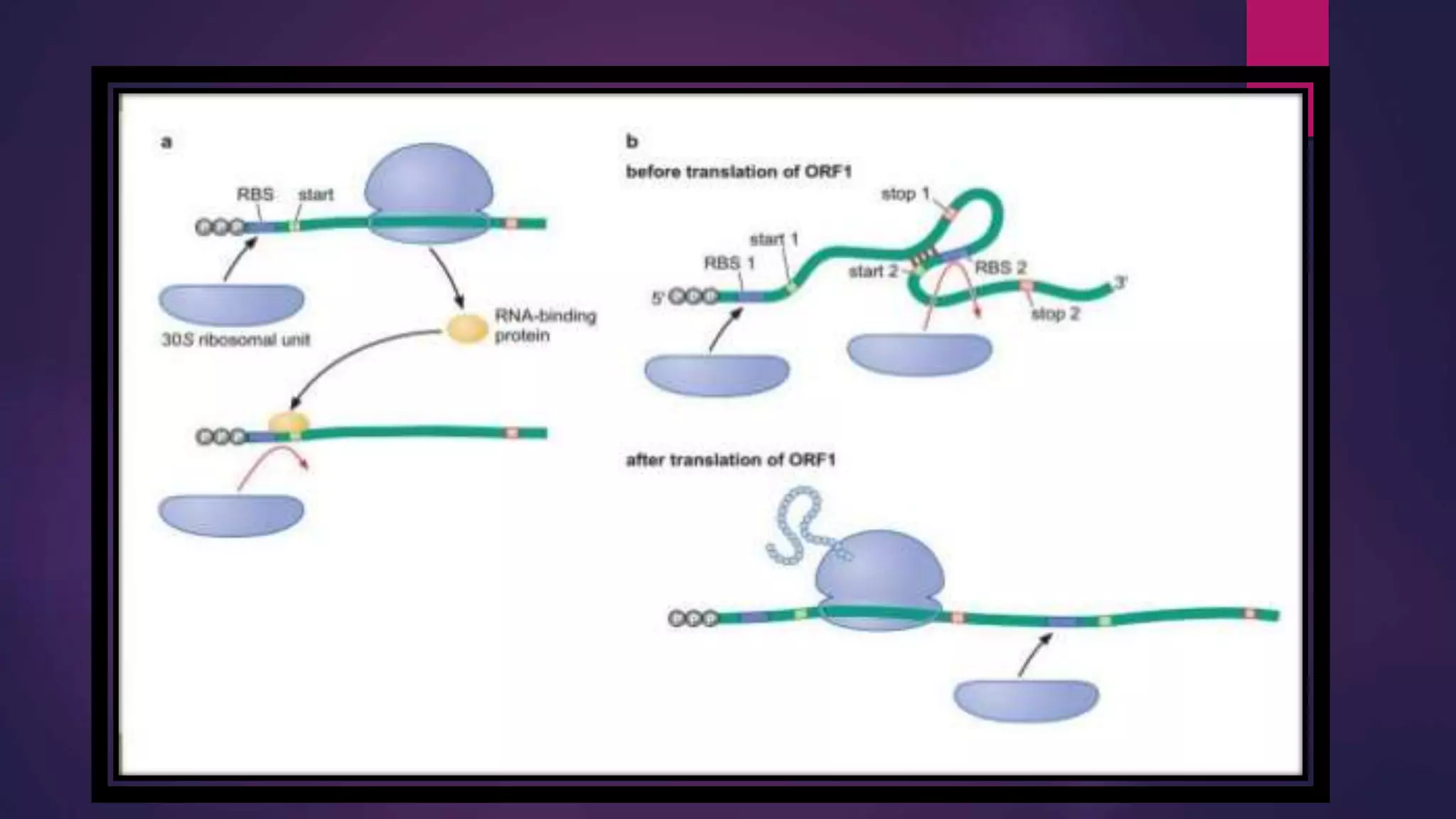
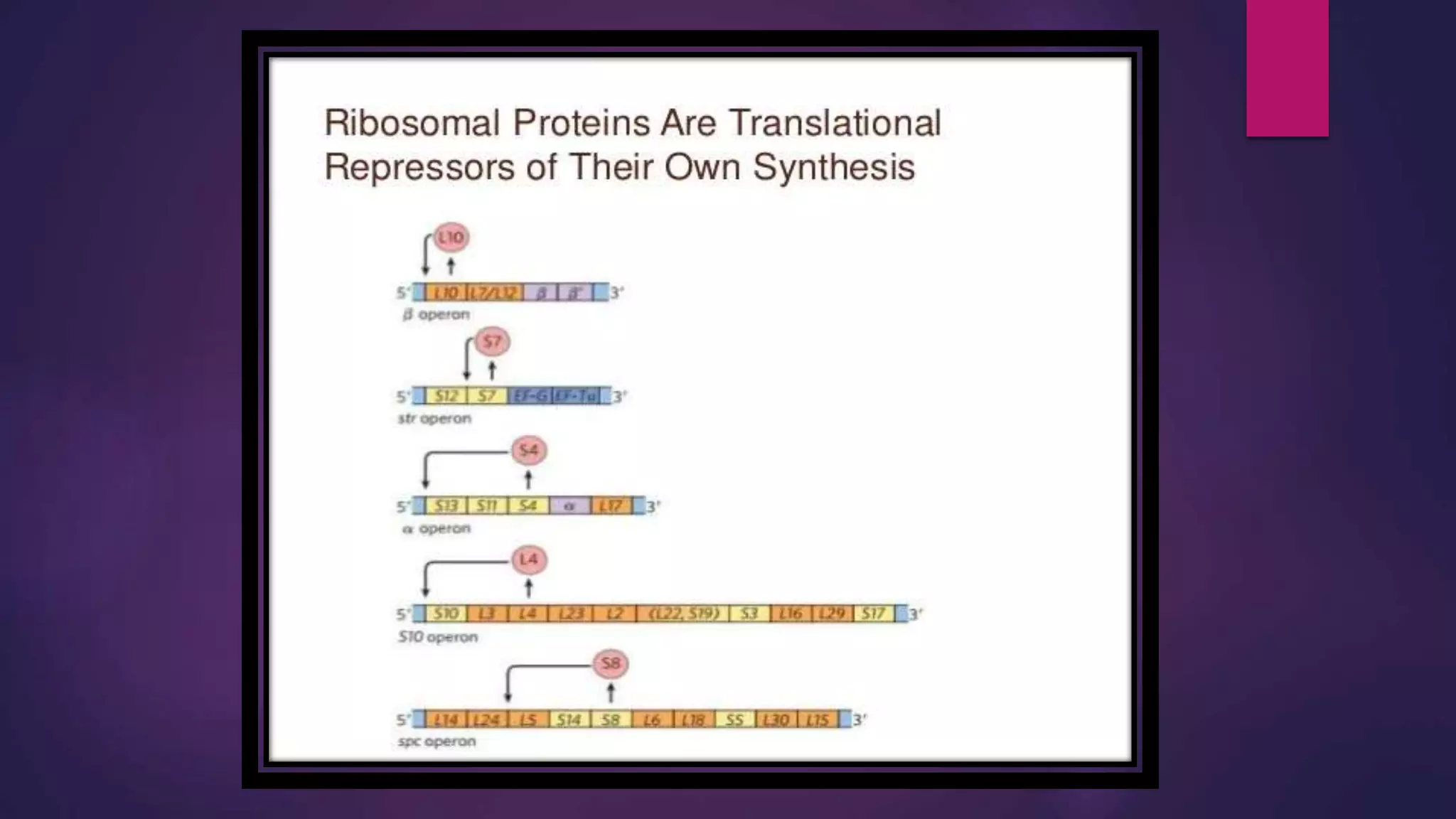
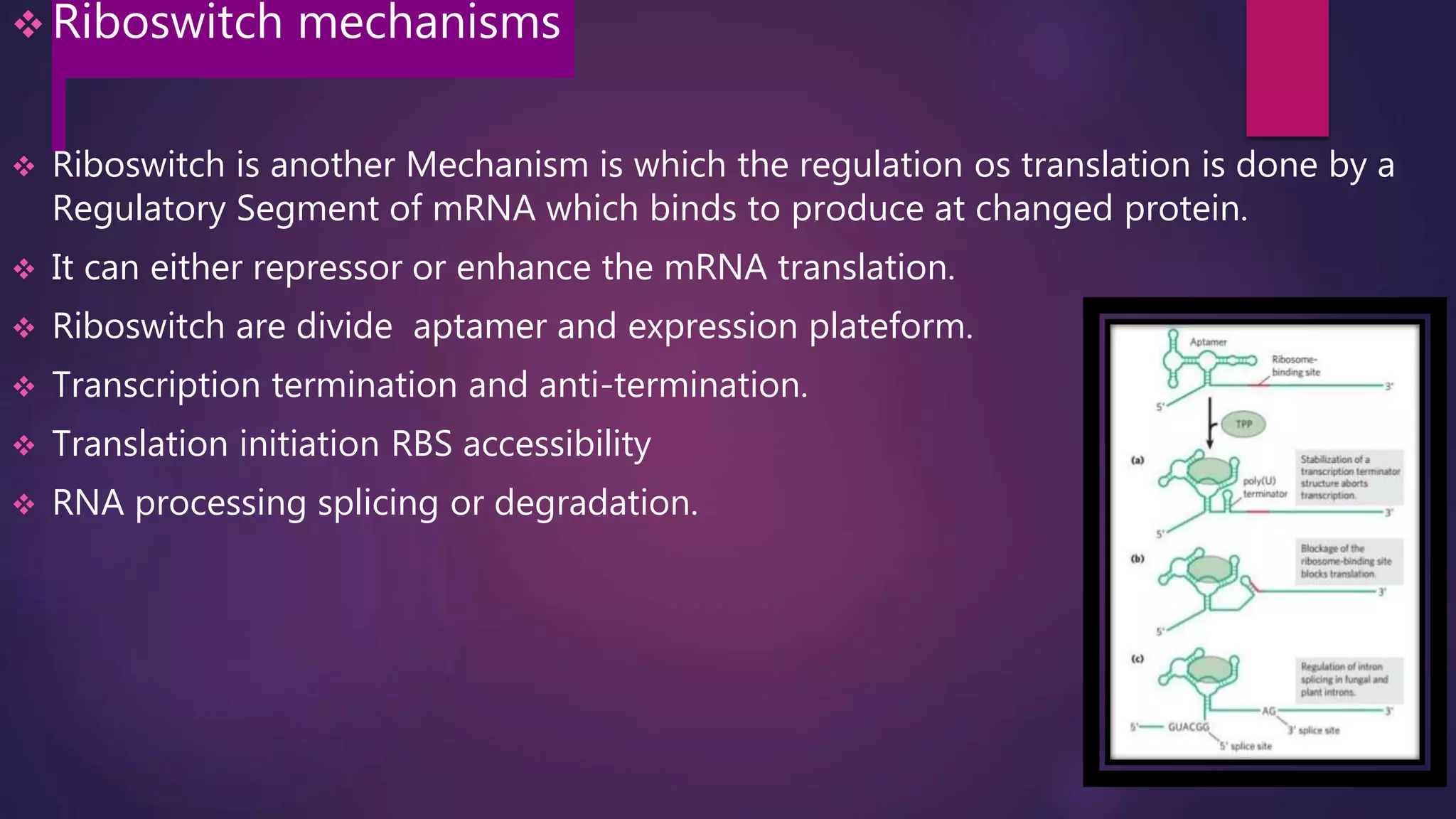
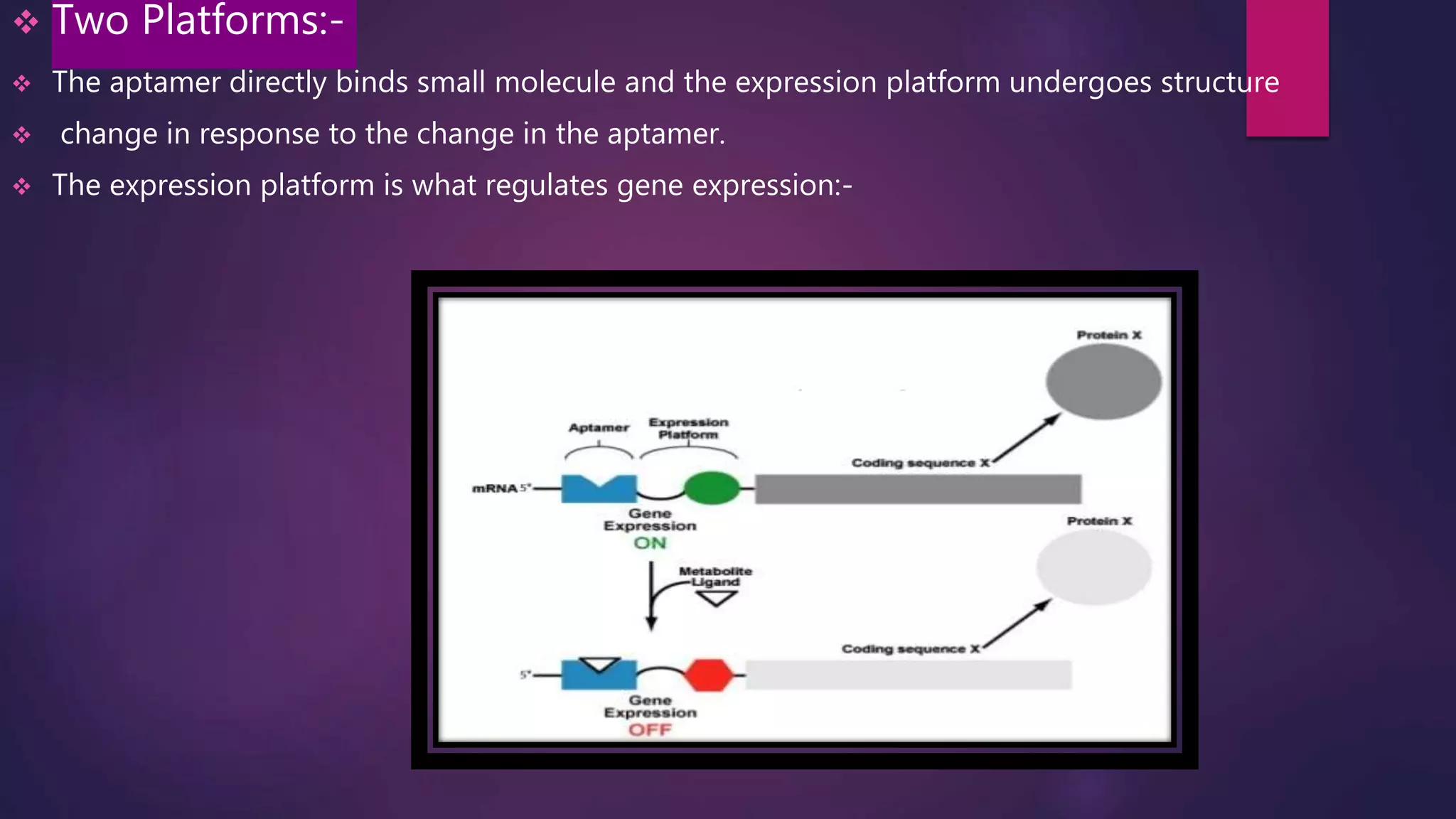
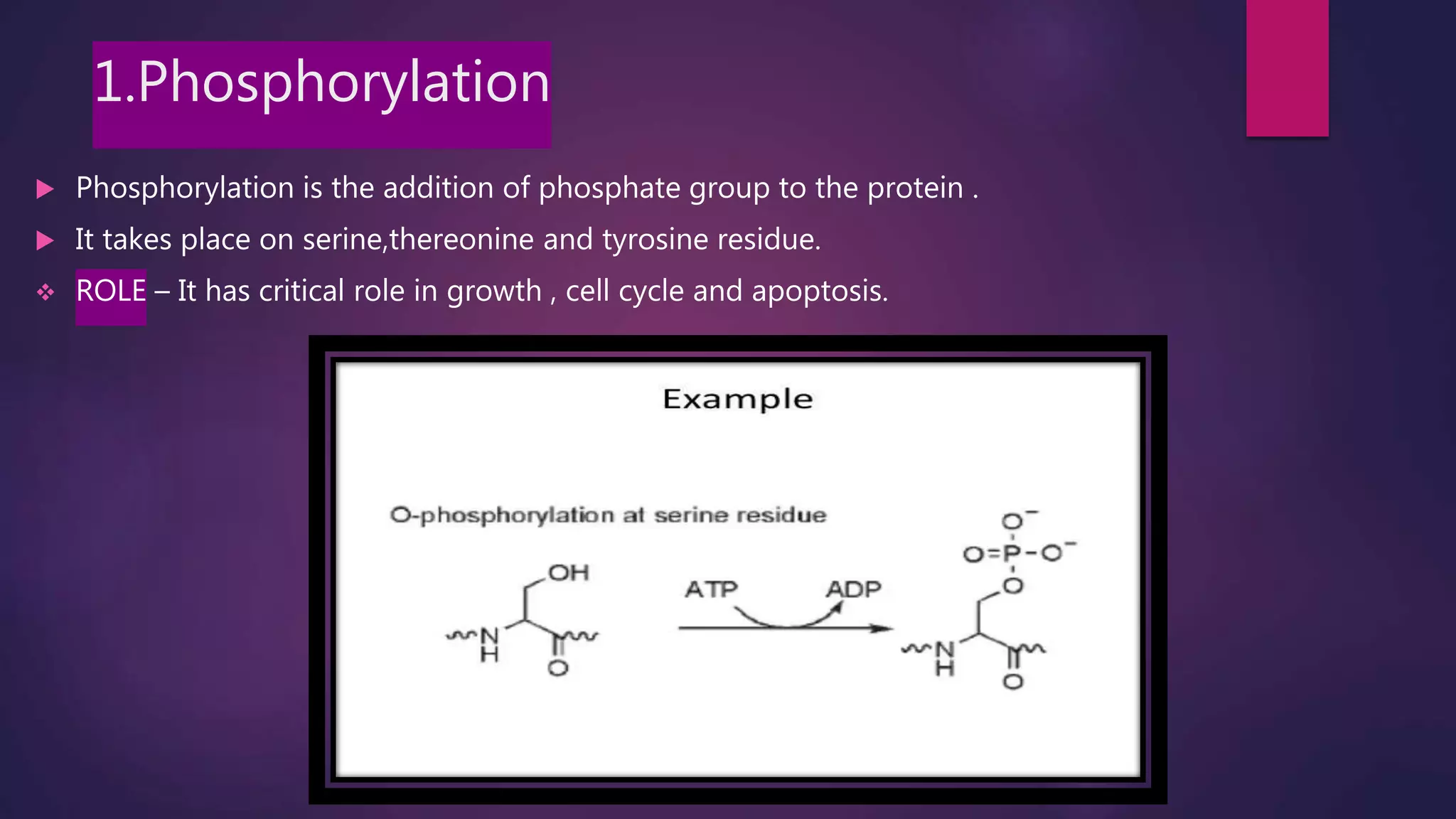
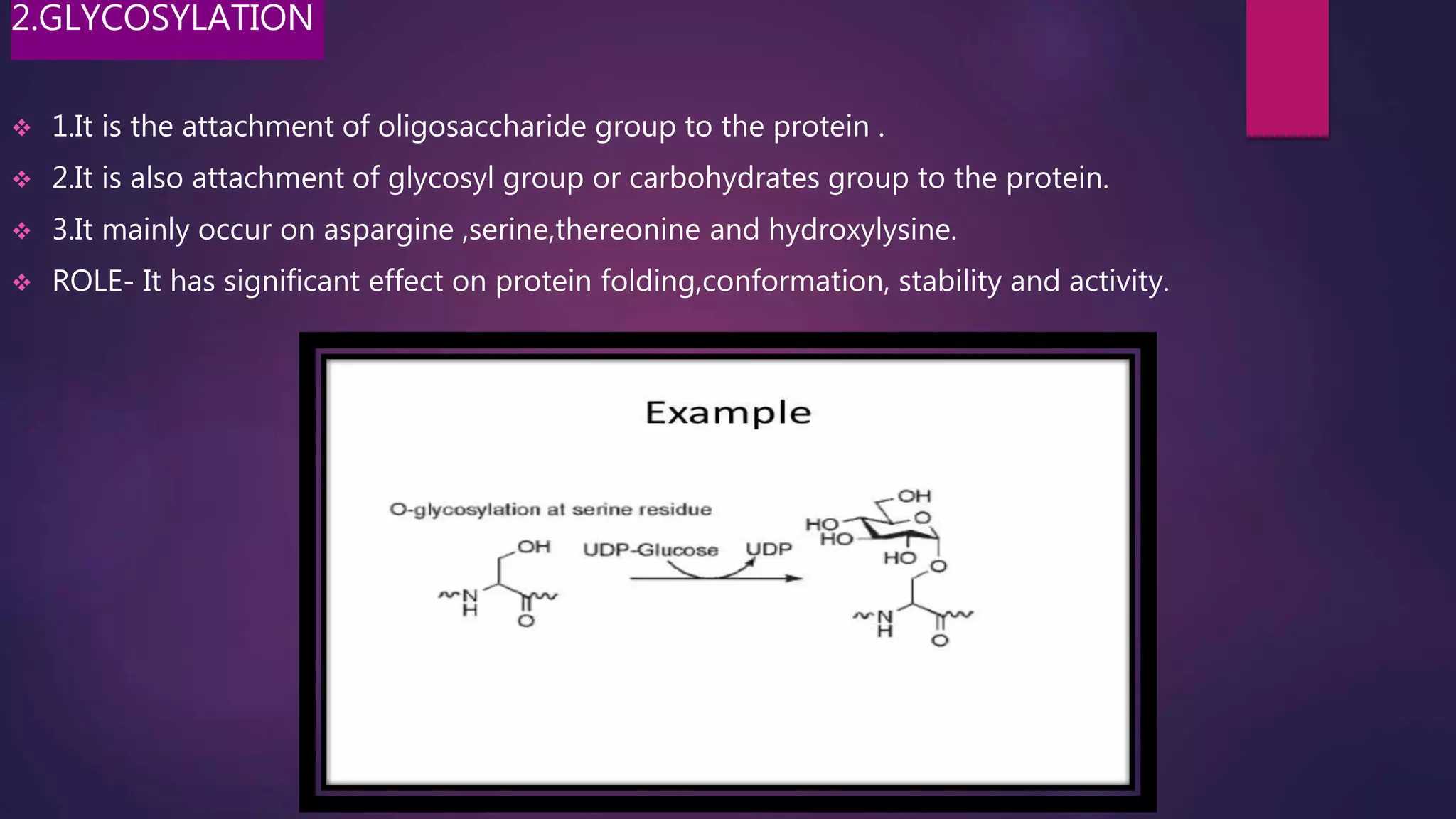
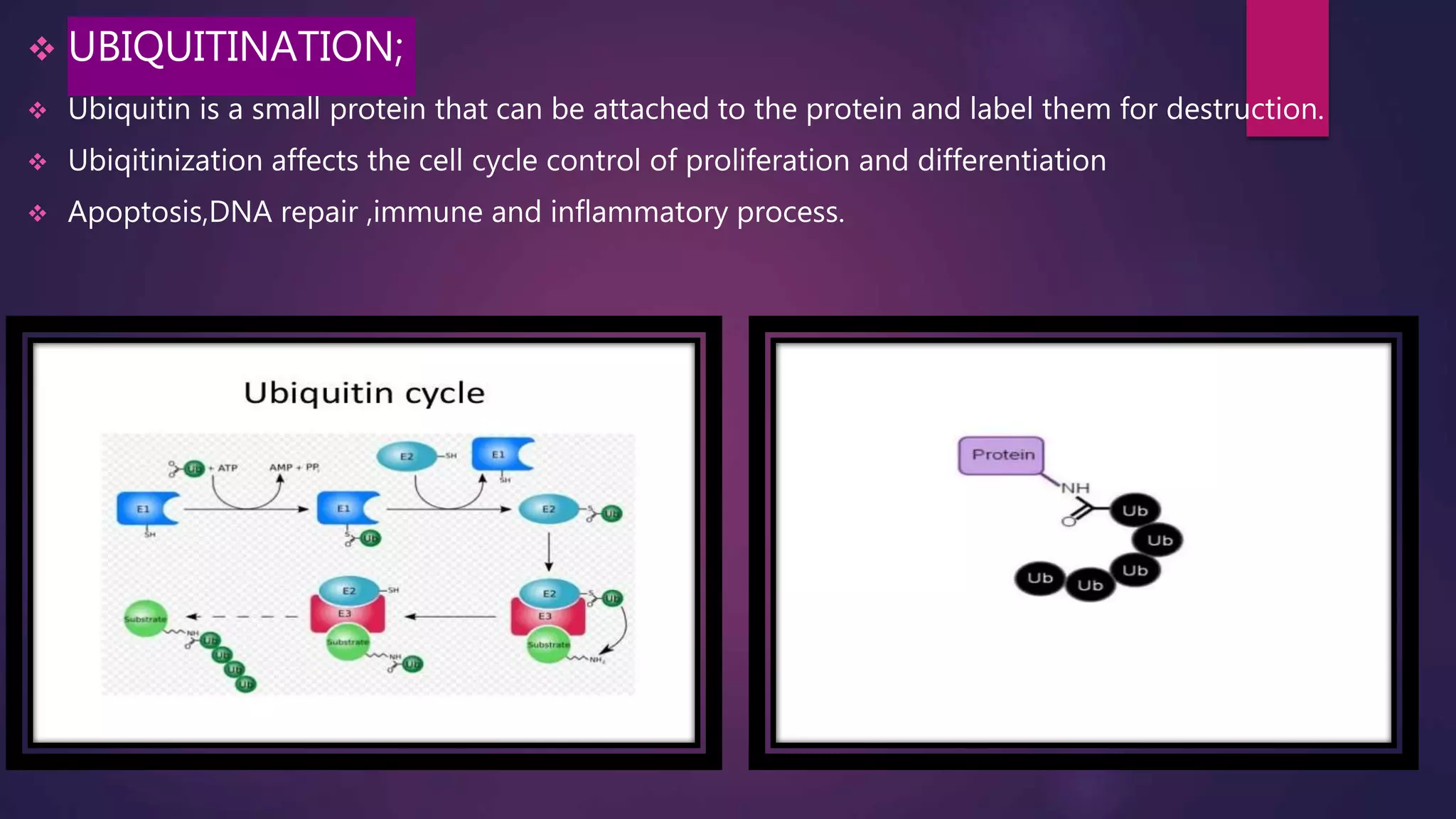
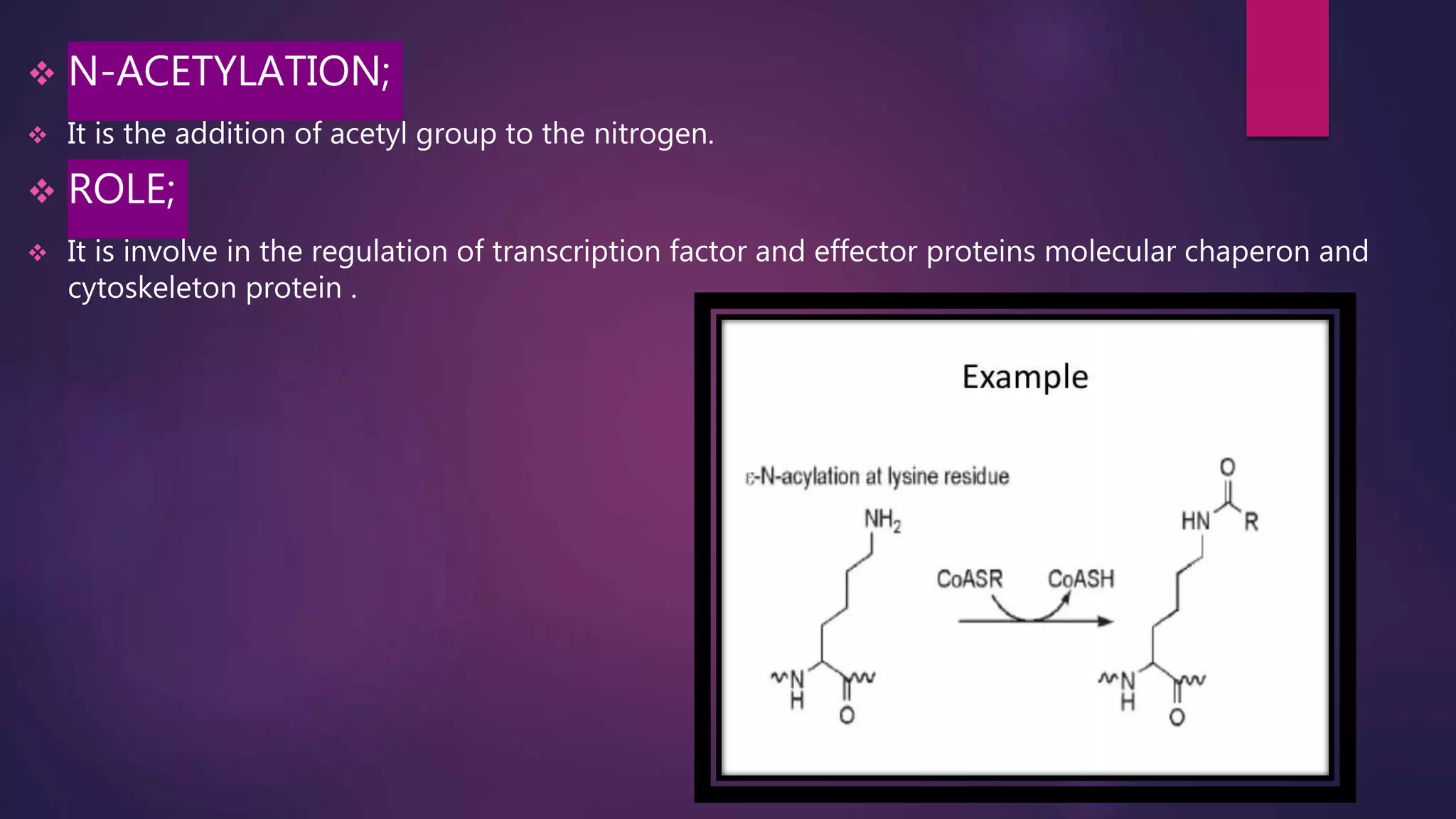
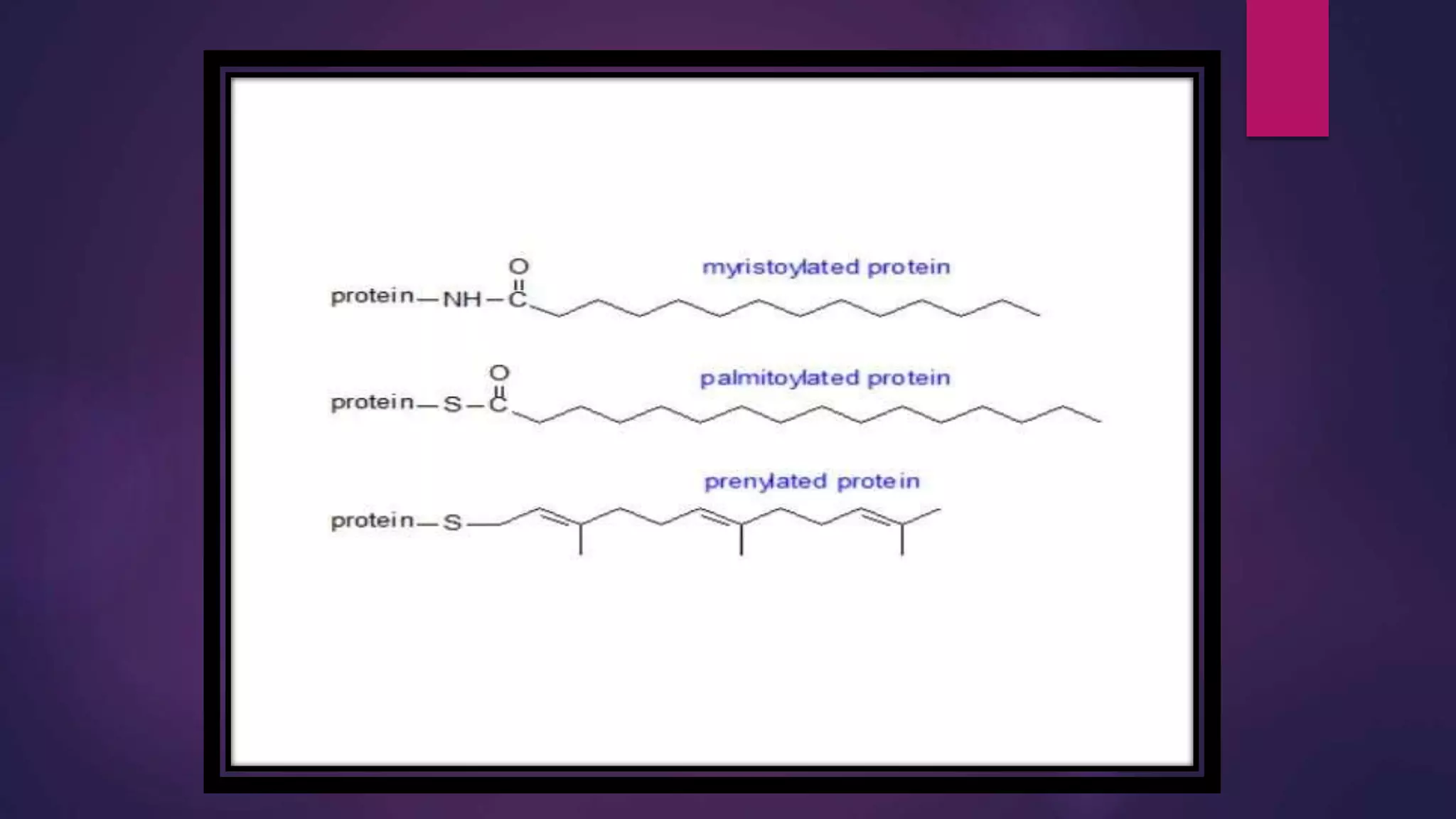
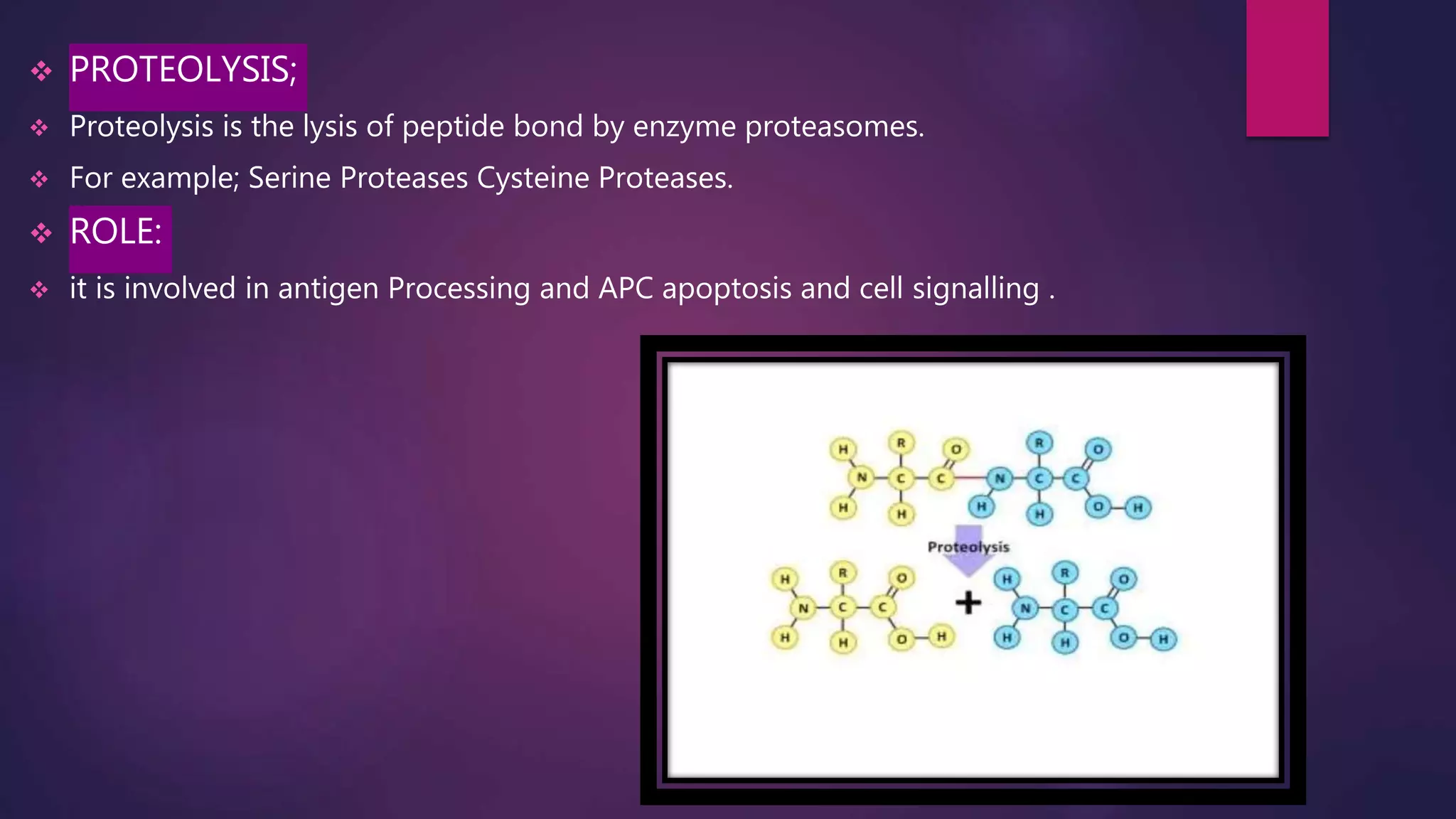
The document provides an extensive overview of the process of translation in both prokaryotes and eukaryotes, detailing the components involved, stages such as initiation, elongation, and termination, and mechanisms of regulation. It discusses the significance of ribosomes, mRNA, and tRNA, along with post-translational modifications like phosphorylation and glycosylation. Key terms, various factors, and techniques for studying translation modifications are also outlined.