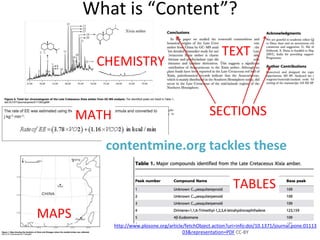
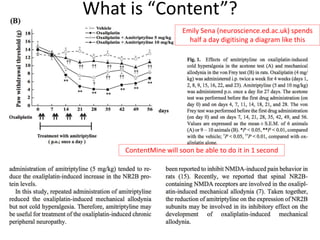
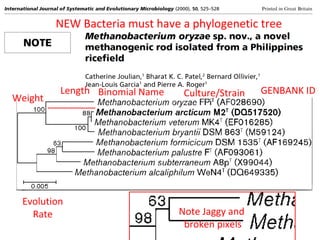
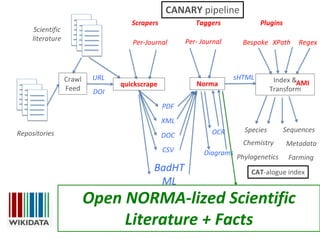
The document discusses the progress and opportunities in content mining for science since the implementation of the Hargreaves legislation in the UK, aiming to make 100 million facts from STEM literature accessible and reusable. It highlights the scale of the task involved, the workshops and collaborations undertaken, as well as the proposed methodologies for extracting and indexing scientific information. The authors emphasize the importance of leveraging content mining to improve research practices and healthcare information sharing in the UK and Europe.



![WHY?
http://www.nytimes.com/2015/04/08/opinion/yes-we-were-warned-about-
ebola.html
We were stunned recently when we stumbled across an article by European
researchers in Annals of Virology [1982]: “The results seem to indicate that
Liberia has to be included in the Ebola virus endemic zone.” In the future,
the authors asserted, “medical personnel in Liberian health centers should be
aware of the possibility that they may come across active cases and thus be
prepared to avoid nosocomial epidemics,” referring to hospital-acquired
infection.
Adage in public health: “The road to inaction is paved with research papers.”
Bernice Dahn is the chief medical officer of Liberia’s Ministry of Health,
where Vera Mussah is the director of county health services. Cameron Nutt
is the Ebola response adviser to Partners in Health.](https://image.slidesharecdn.com/wtfinal-150415071510-conversion-gate01/85/Content-Mining-at-Wellcome-Trust-4-320.jpg)