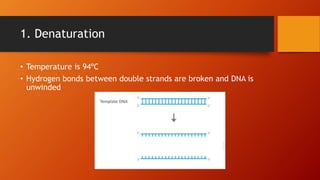
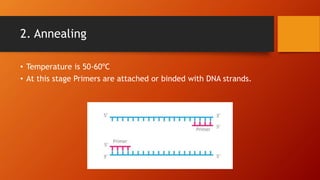
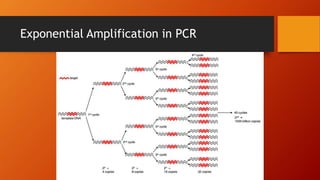
Polymerase chain reaction (PCR) is a technique used to amplify DNA sequences. It involves repeated cycles of heating and cooling of the DNA sample to cause DNA replication. Each cycle doubles the number of copies of the target DNA sequence. Key steps in PCR include DNA denaturation to separate strands, annealing of primers to the strands, and elongation of new strands by a polymerase enzyme. Factors like primer concentration, temperature, and number of cycles influence the exponential amplification of the target DNA sequence. PCR has many applications in research, forensics, and medicine.