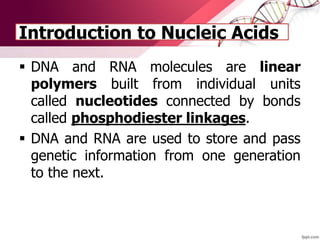
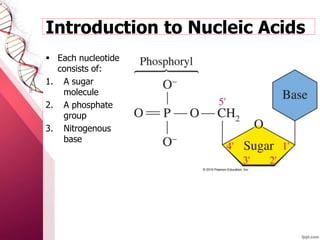
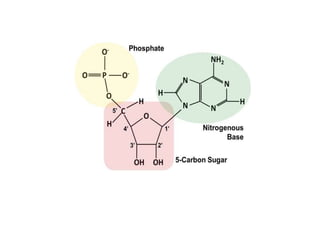
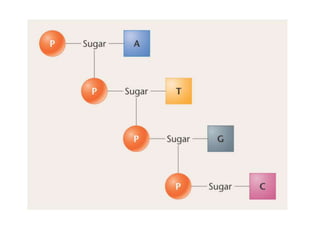
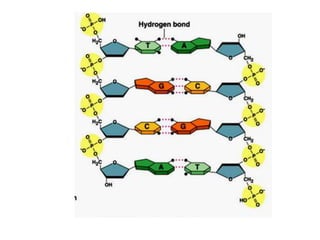
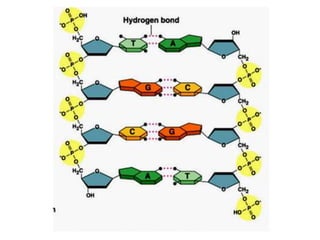
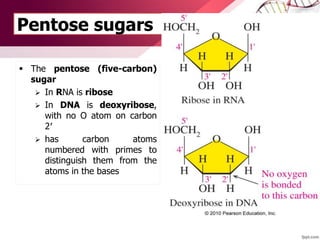
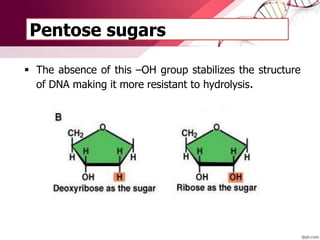
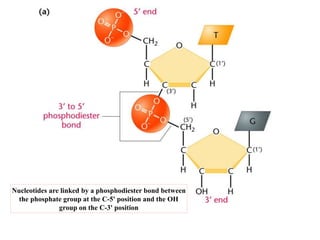
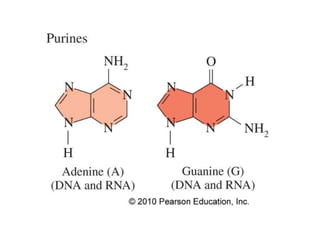
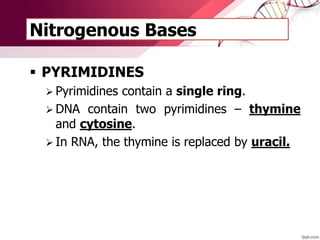
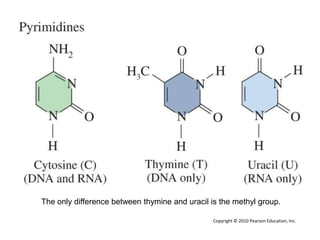
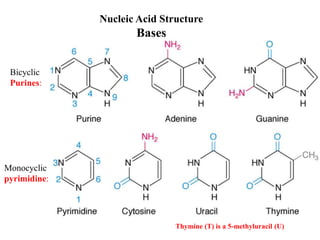
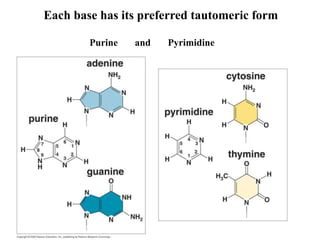
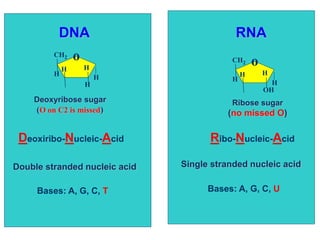
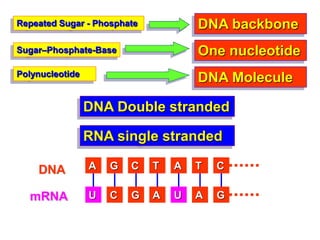
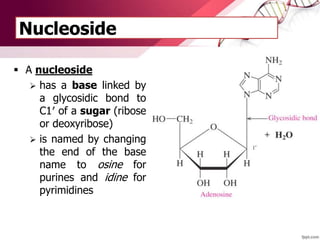
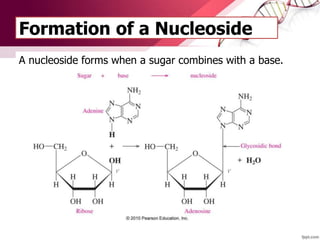
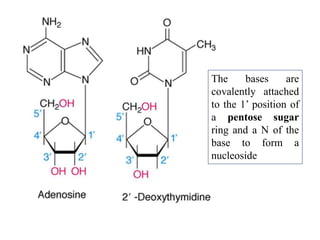
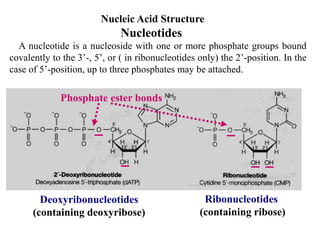
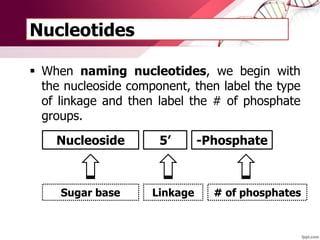
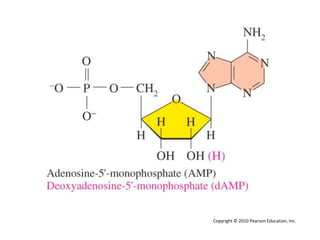
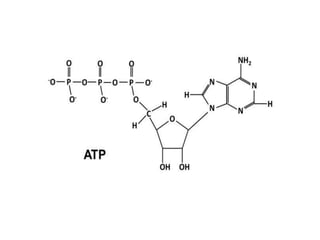
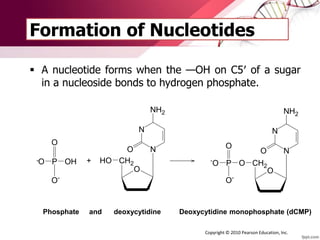
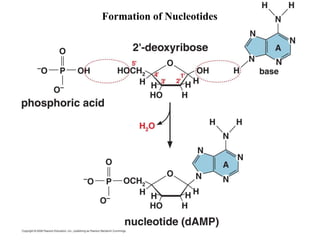
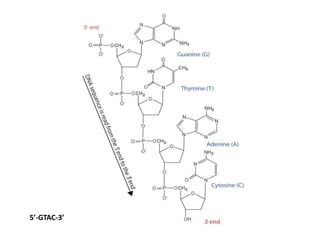
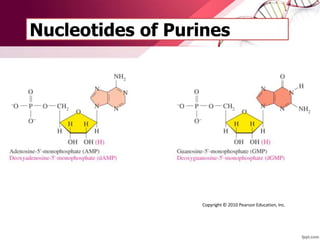
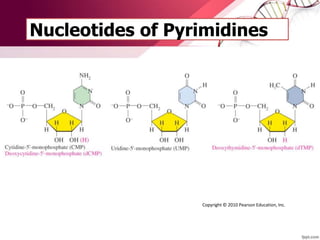
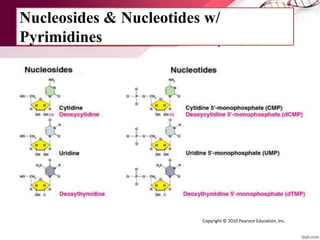
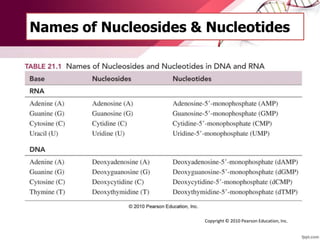
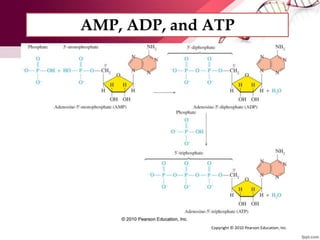
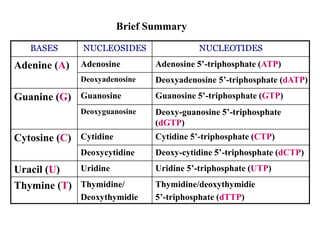
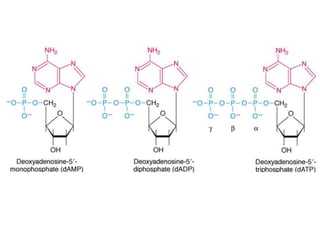
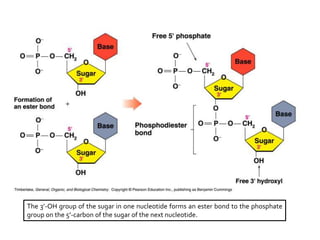
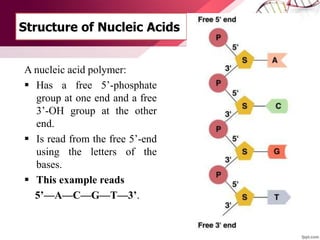
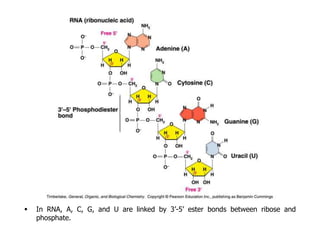
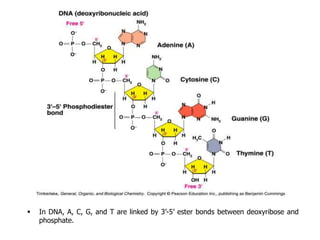
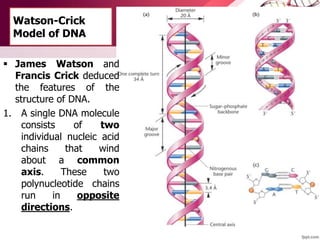
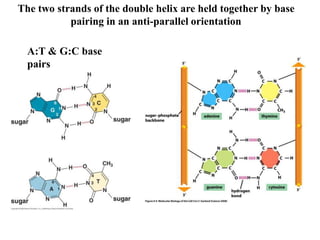
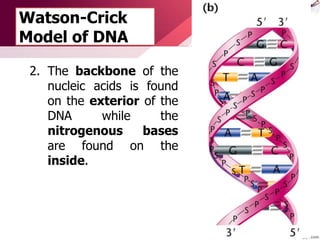
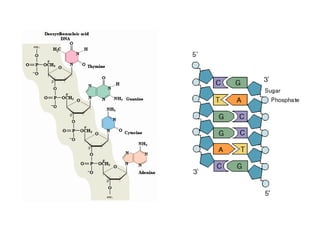
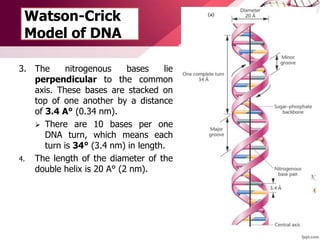
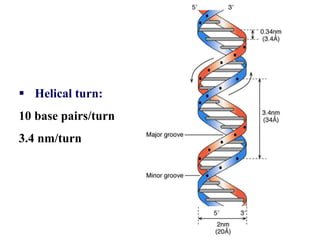
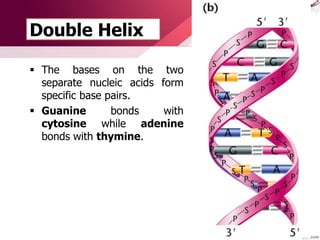
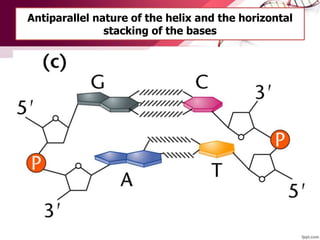
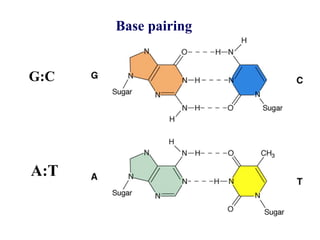
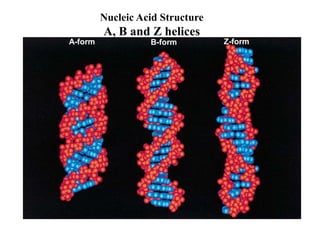
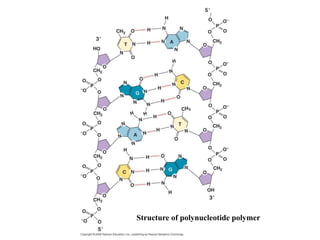
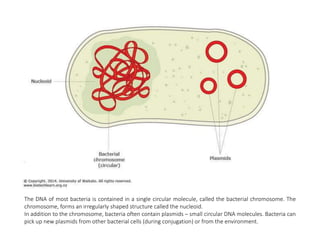
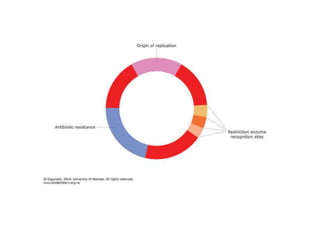
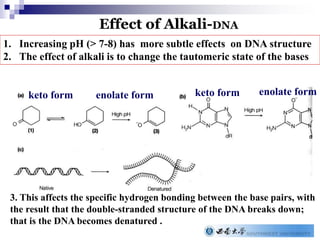
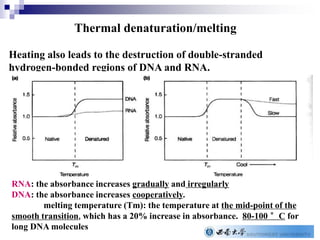
The document provides an introduction to nucleic acids, detailing their structure, types, and functions. It explains that DNA and RNA are both composed of nucleotides, each containing a sugar, phosphate group, and nitrogenous base, with DNA forming a double helix and RNA typically existing as a single strand. Additionally, it covers the roles of nucleic acids in genetic information storage and protein synthesis, as well as the chemical properties that affect their stability.