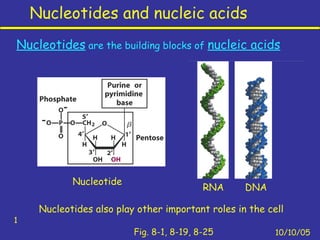
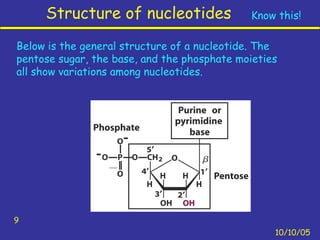
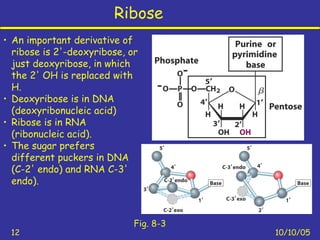
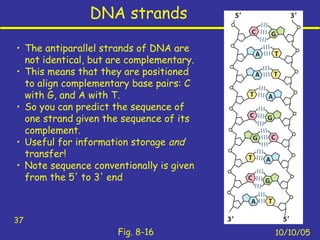
Nucleotides are the building blocks of nucleic acids and consist of a nitrogenous base, a pentose sugar, and a phosphate group. Nucleotides play key roles as the monomers of nucleic acids DNA and RNA, as well as functioning as energy carriers like ATP and cofactors like NAD. Nucleic acids such as DNA contain the genetic information through base pairing and various structures enable different functions like protein synthesis and catalysis.