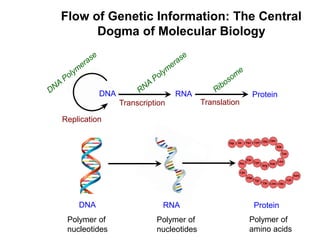
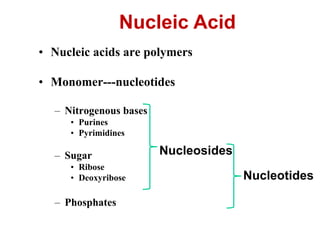
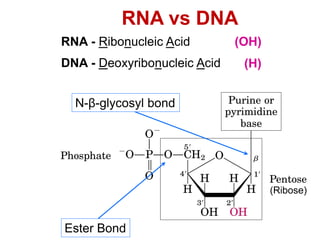
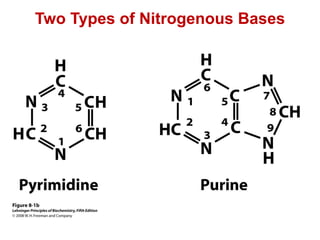
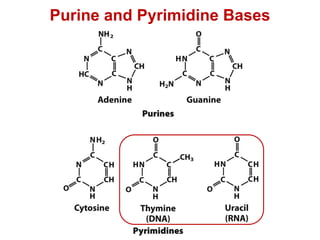
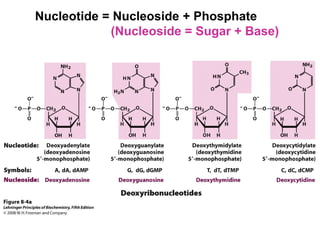
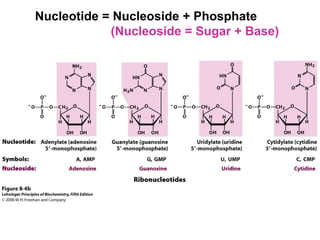
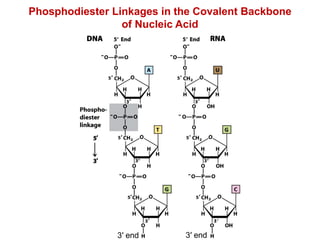
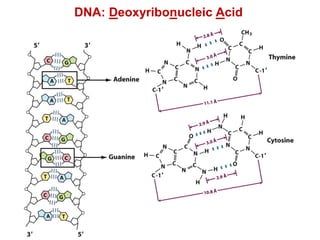
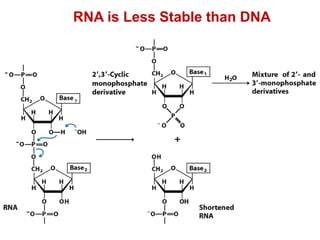
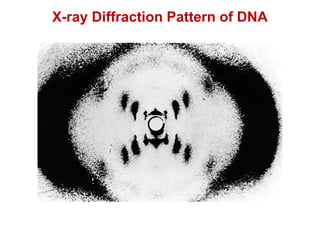
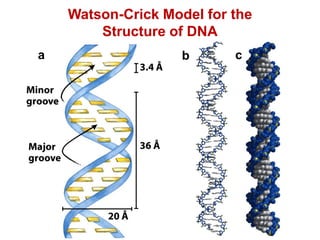
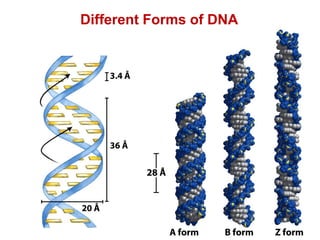
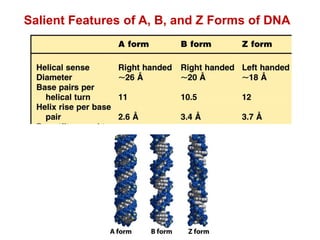
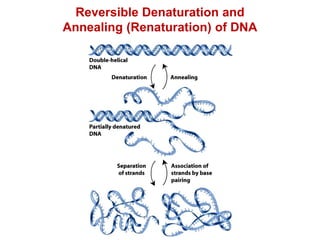
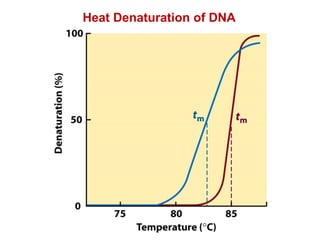
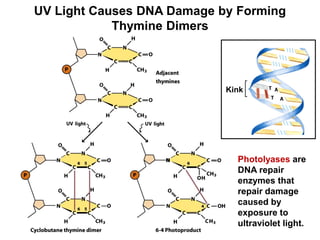
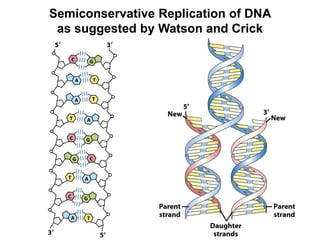
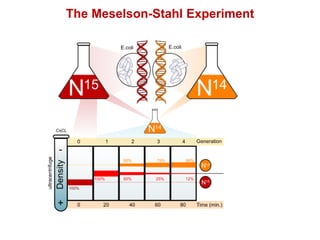
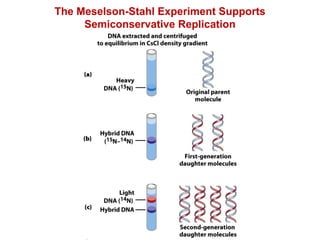
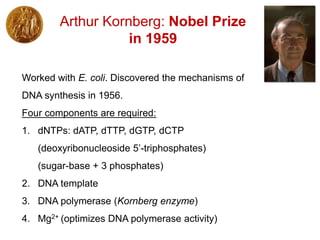
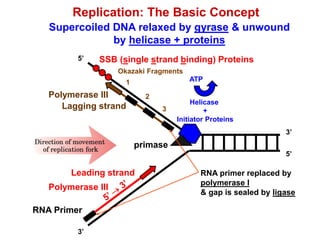
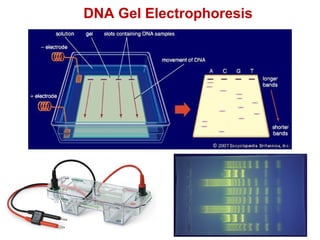
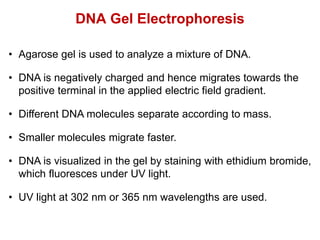
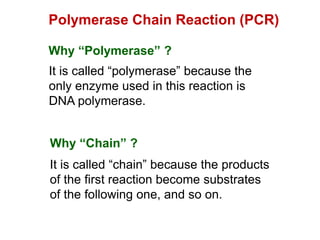
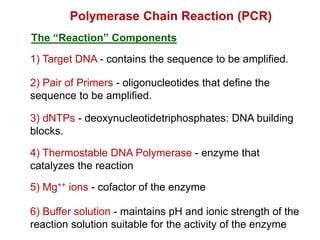
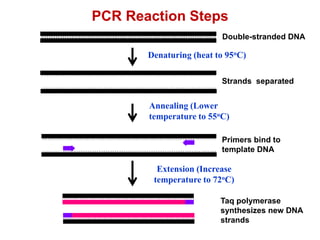
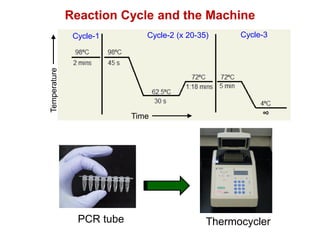
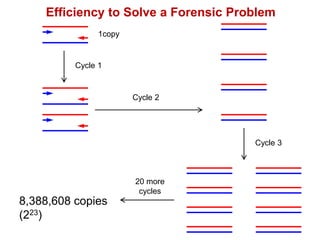
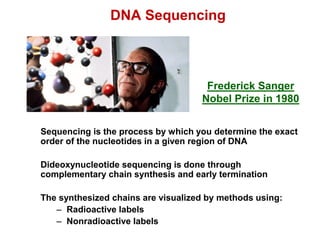
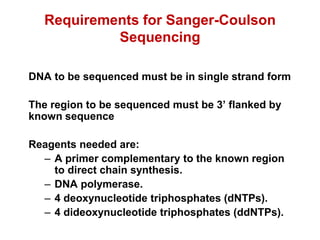
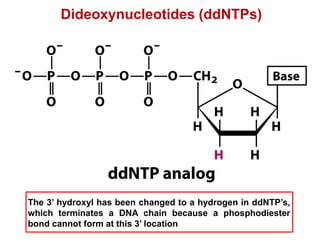
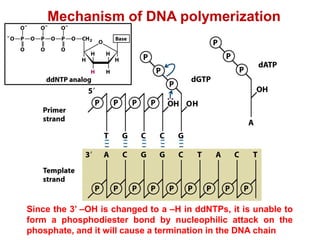
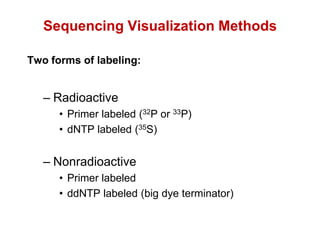
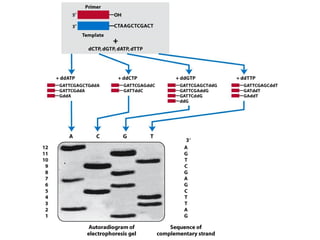
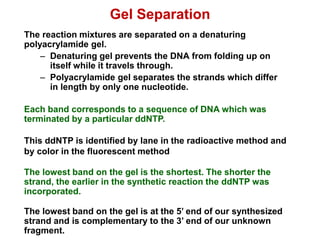
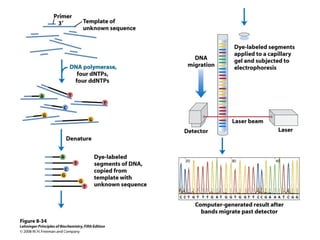
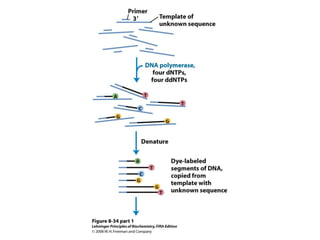
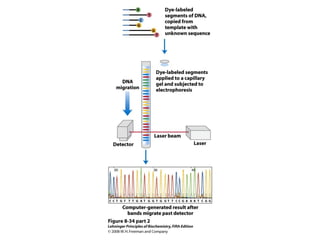
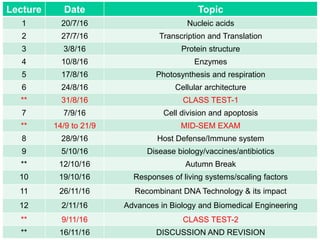
This document outlines the schedule and topics for a course on the science of living systems. The course covers topics such as nucleic acids, transcription and translation, protein structure, enzymes, photosynthesis and respiration, cell division, the immune system, and recombinant DNA technology. It includes the dates for lectures, exams, and breaks. The document also provides background information on water and its importance for life, as well as DNA structure and replication. Key concepts in molecular biology such as the central dogma are explained.



![Henderson–Hasselbalch
Equation
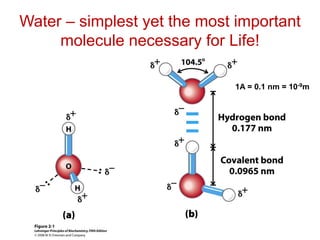
Water is ionizable
H2O + H2O = H3O+ + OH-
KW (equilibrium constant)
[H+] x [OH-]
[H2O]
=
HA = H+ + A-
Ka (equilibrium constant)
[H+] x [A-]
[HA]
=
pH = pKa + log10
[A-]
[HA]](https://image.slidesharecdn.com/lecture1nucleicacidemailed-170211180536/85/Nucleic-acid-9-320.jpg)