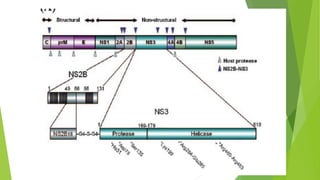
The document discusses the global consensus sequence development and analysis of conserved domains in the NS3 protein of dengue virus. It describes how over 125 sequences of the NS3 protein from the four dengue virus serotypes were aligned and analyzed. Key conserved domains that encode the serine protease and helicase functions were identified. The phylogenetic tree analysis grouped the sequences by serotype. Suggestions for potential antiviral peptides targeting conserved motifs in these domains to disrupt protein functions and reduce viral replication are provided.











![Suggestions
The consensus sequence alignment shows that:
1.Residue L226 to L245 encodes the P-loop NTPase superfamily. It has two
motifs [Walker A, GK(S/T), and Walker B, Dex (D/H)], designing antipeptides
against the Walker A/Walker B region can significantly reduce RNA processing
efficiency.
2.I349 to R357 encodes helicase DEAD-box domain. Designing Antipeptides against
DEAD-Box can control Viral activites
3.The peptide string from 465 to 473 (QRR267 GR469 XGRN) is the part of the
helicase domain , mutation in R467 and R469 that will reduce the activity of
helicase
4.Mutation in residues (Y159 and G160) to alanine can completely
abrogate the enzyme activity](https://image.slidesharecdn.com/presentation1inrtoduction-141213053503-conversion-gate01/85/NS2-conserved-Domain-In-Dengue-Virus-22-320.jpg)
