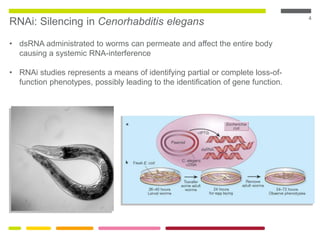
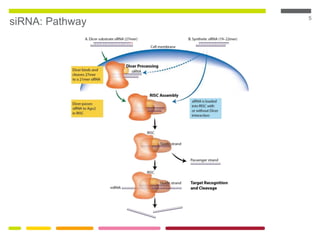
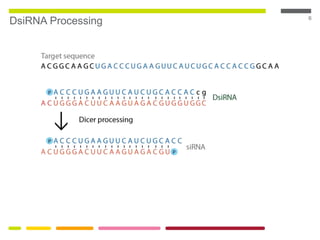
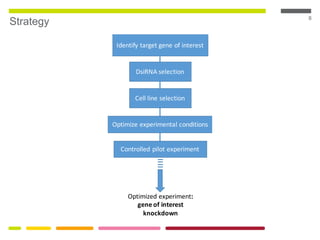
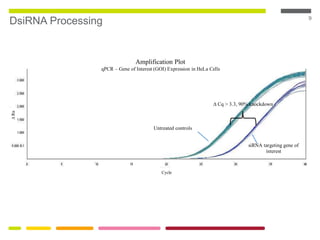
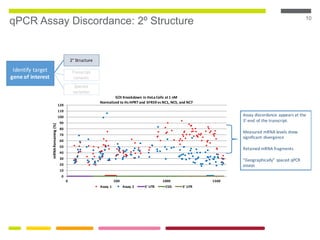
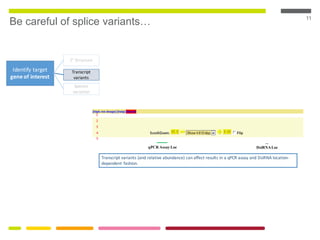
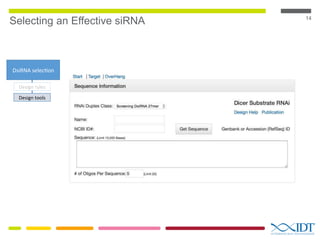
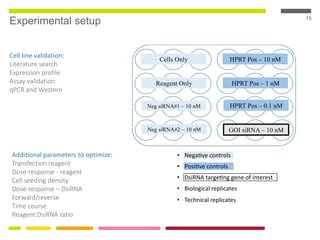
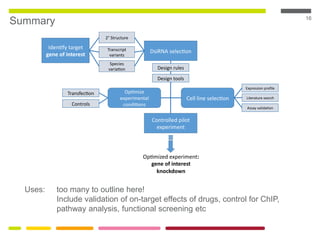
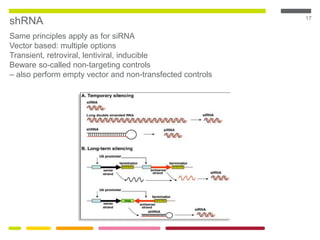
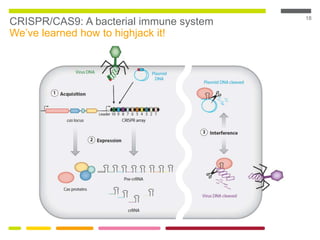
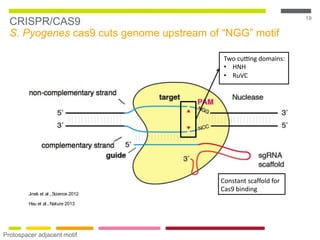
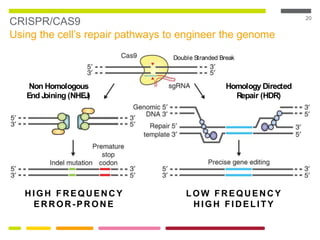
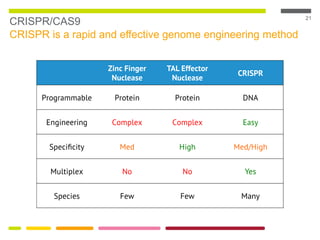
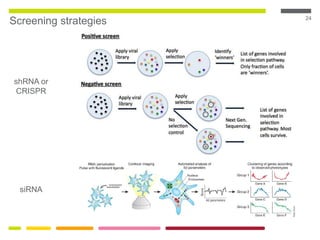
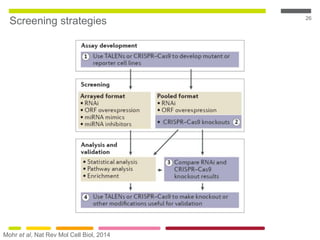
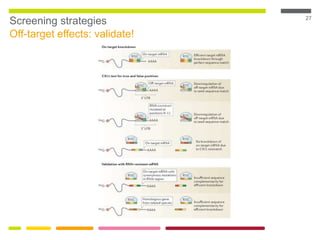
This document discusses RNA interference (RNAi) techniques such as silencing RNA and CRISPR/Cas9. It explains that double-stranded RNA is cut by the enzyme Dicer into short interfering RNAs (siRNAs) that can degrade mRNA strands in a highly specific process. RNAi is involved in regulating 30% of the human genome and acts as a defense mechanism against viruses and transposons. The document also discusses selecting effective siRNAs, considerations for species variation and secondary RNA structures, and strategies for gene knockdown screening using shRNA and CRISPR/Cas9.