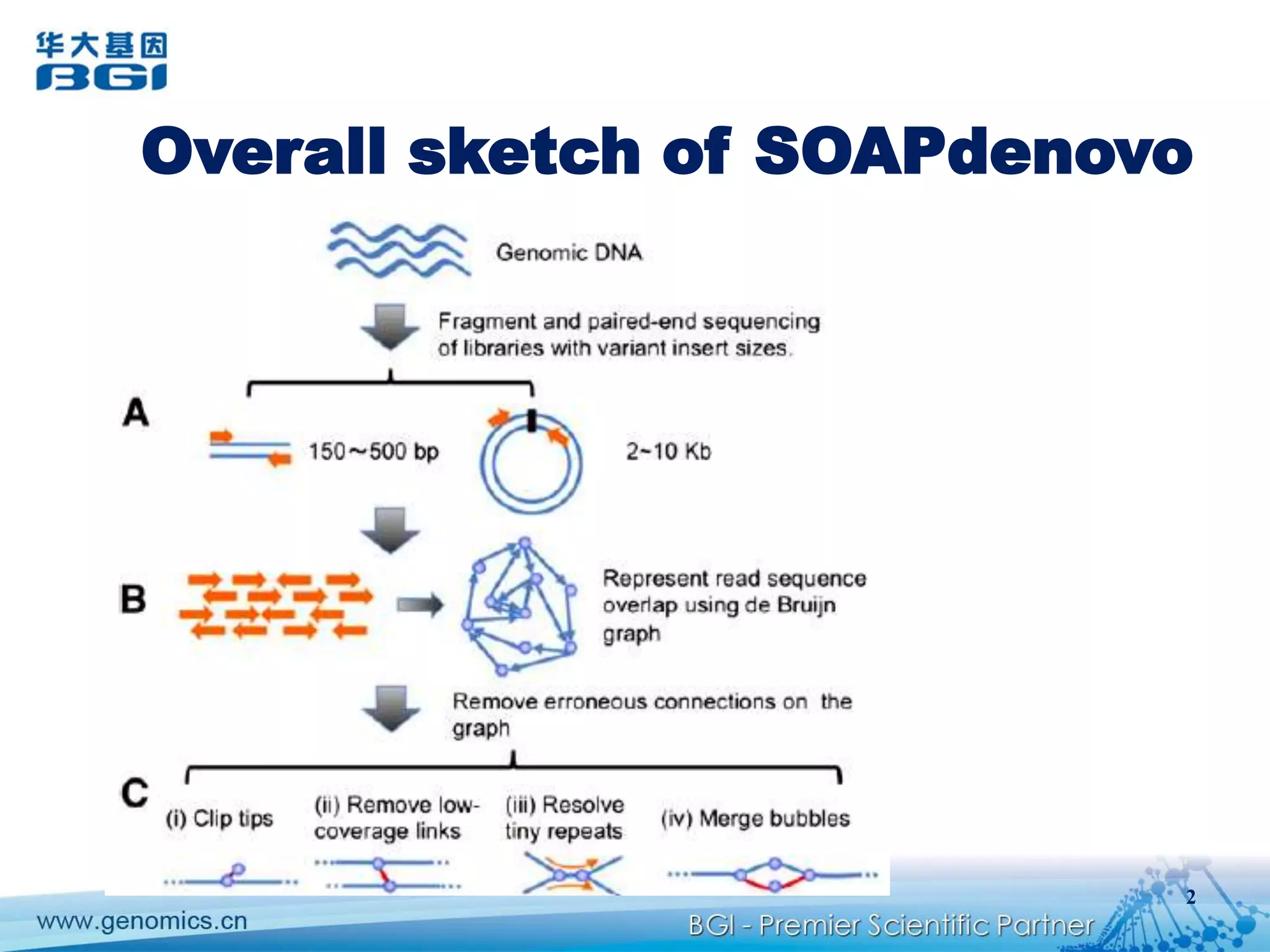
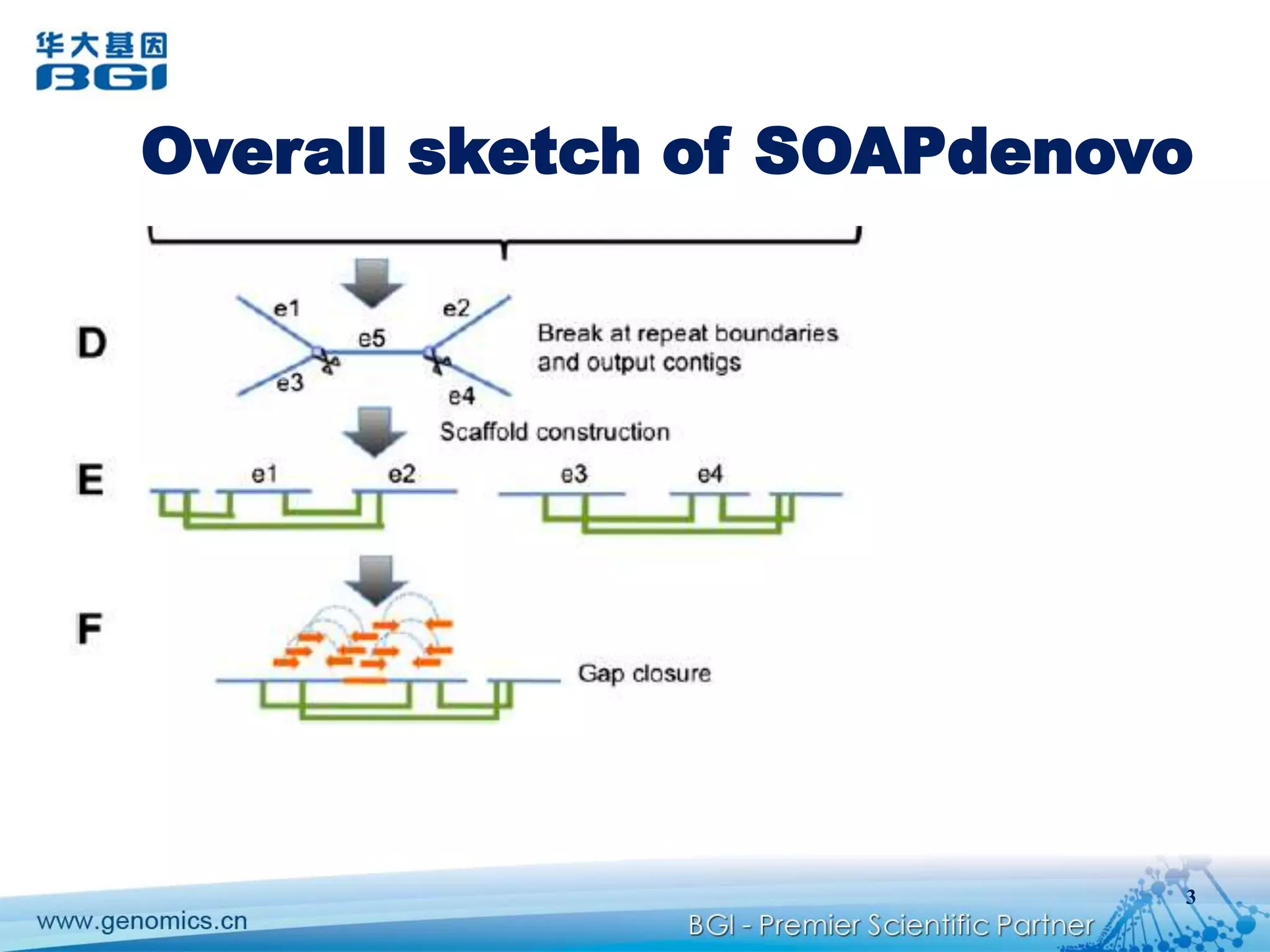
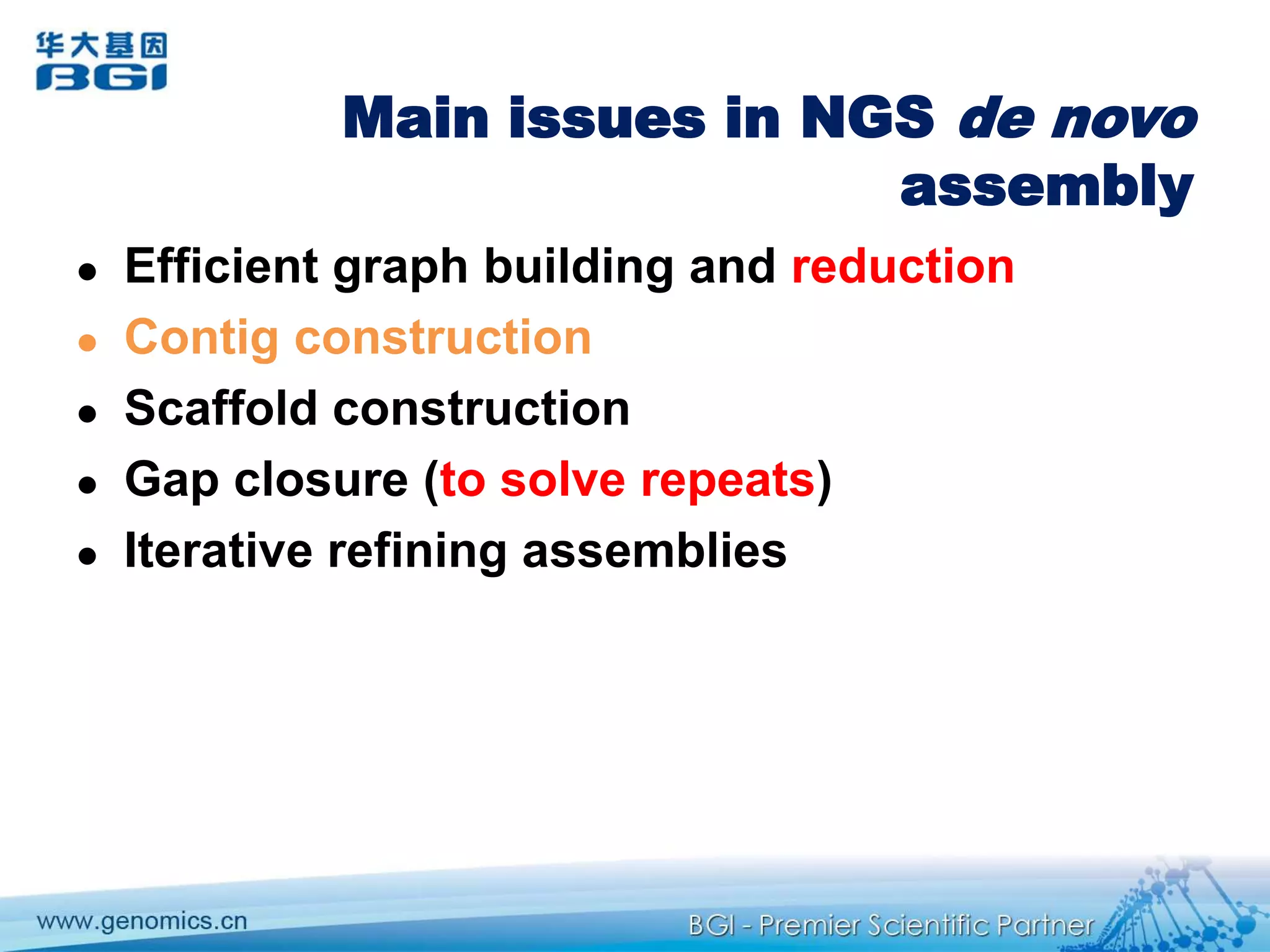
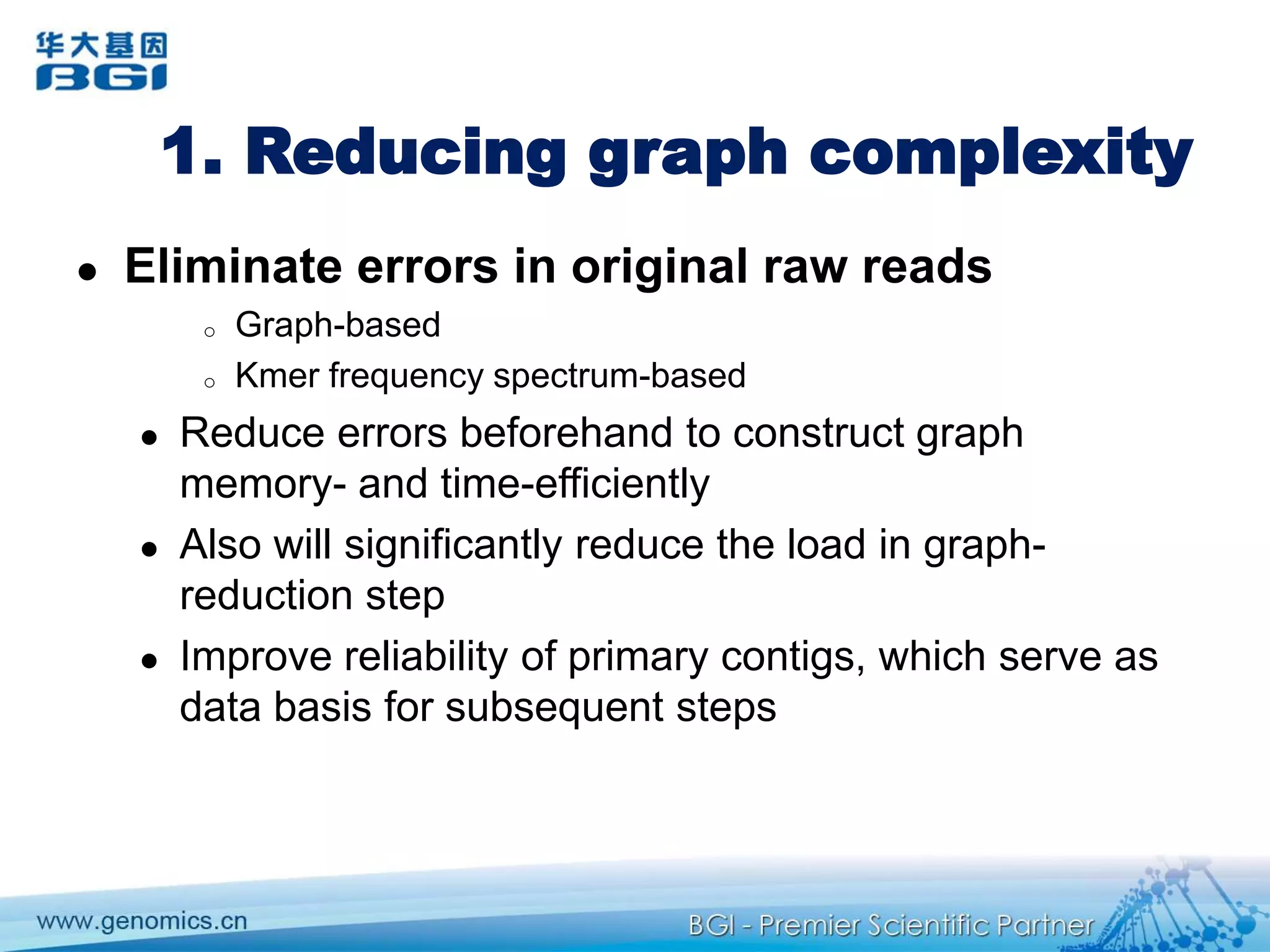
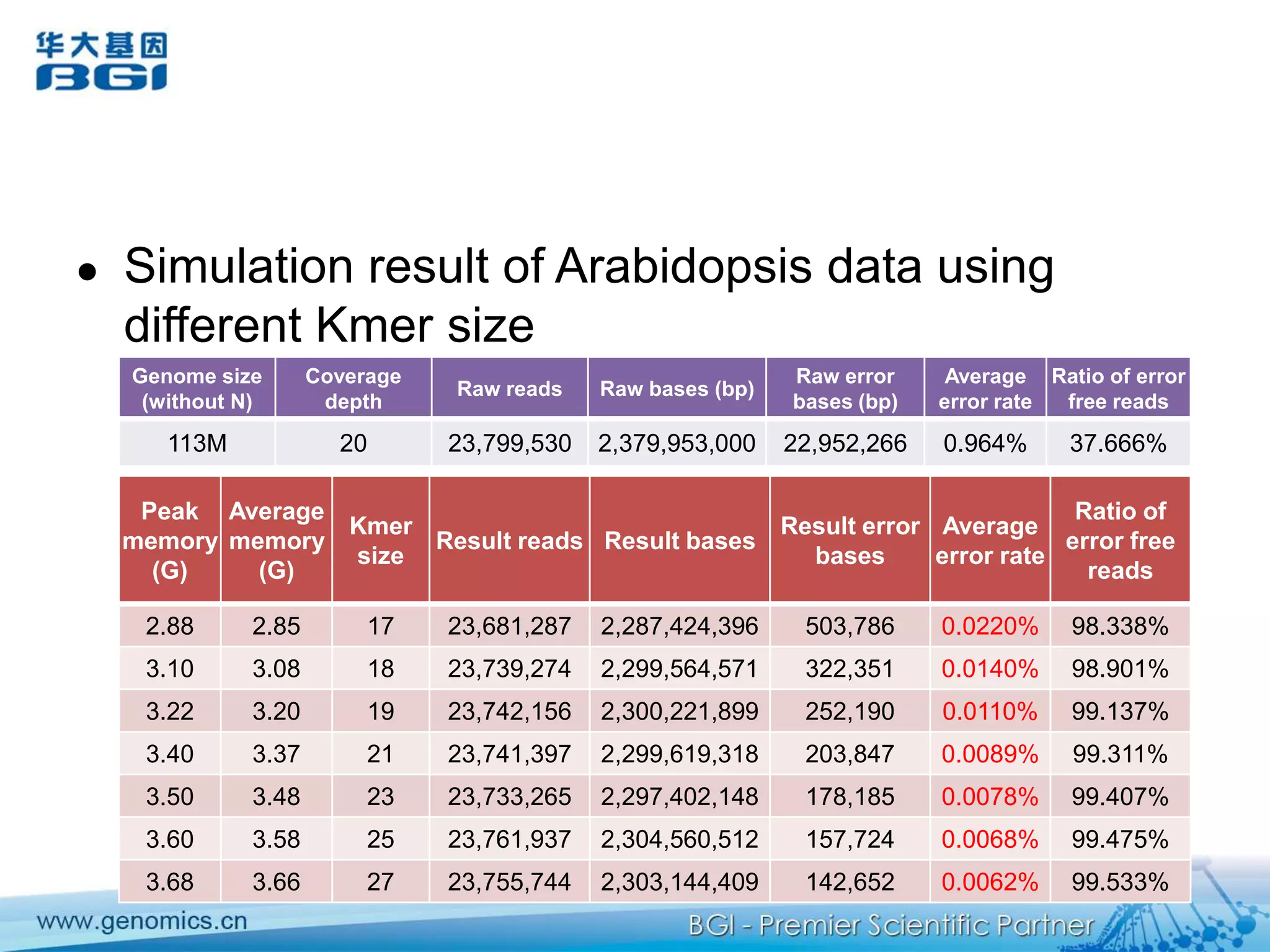
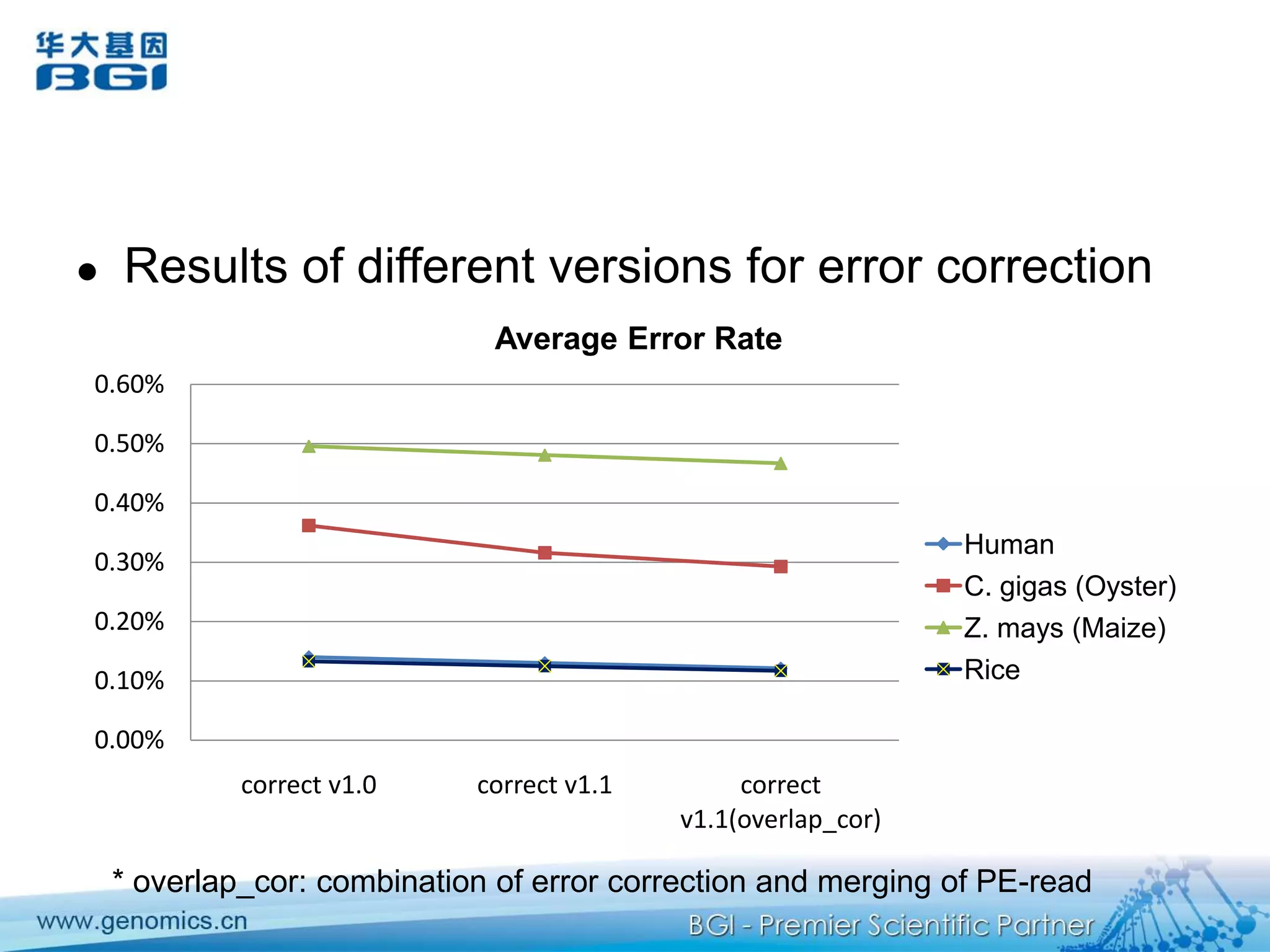
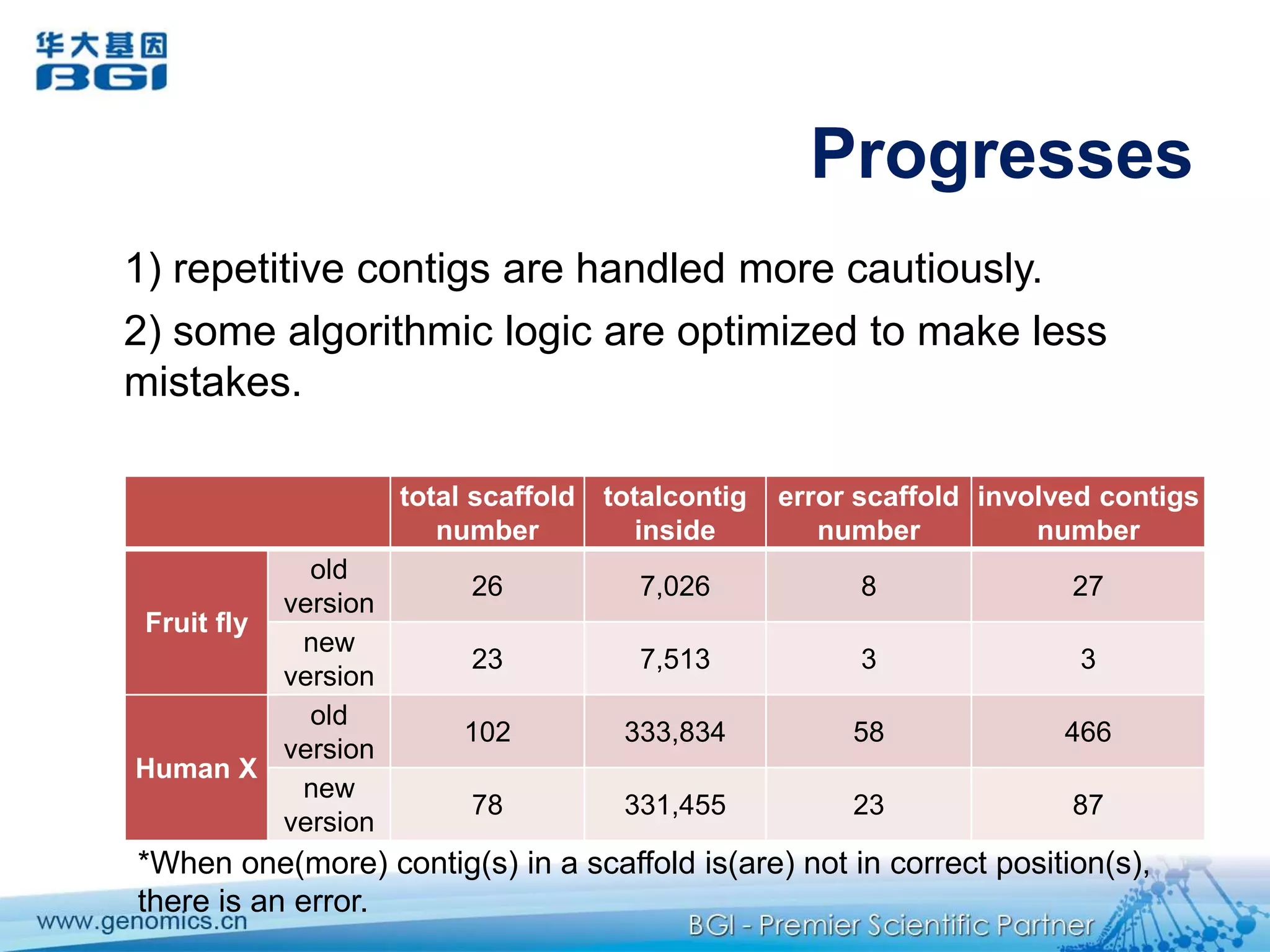
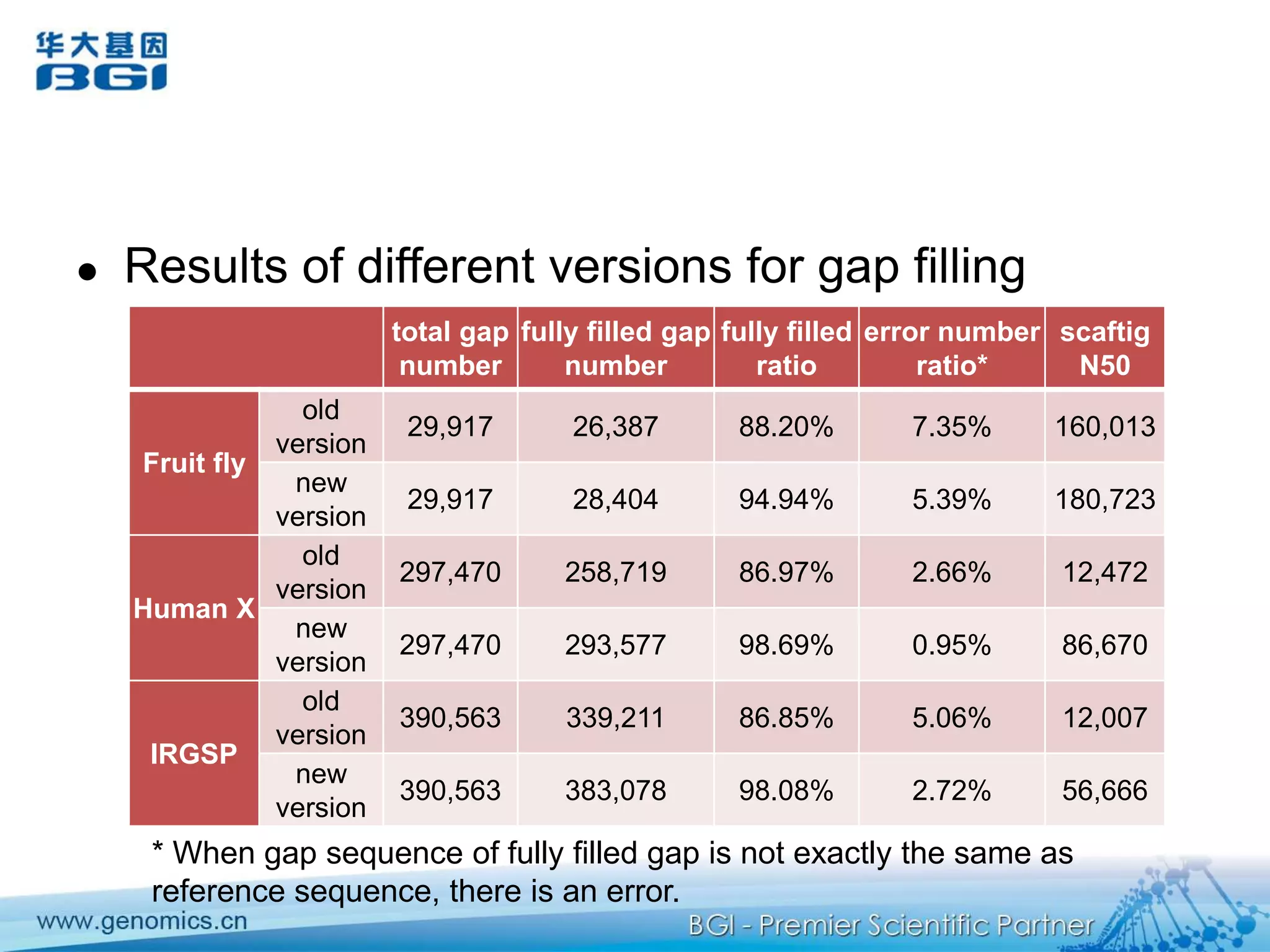
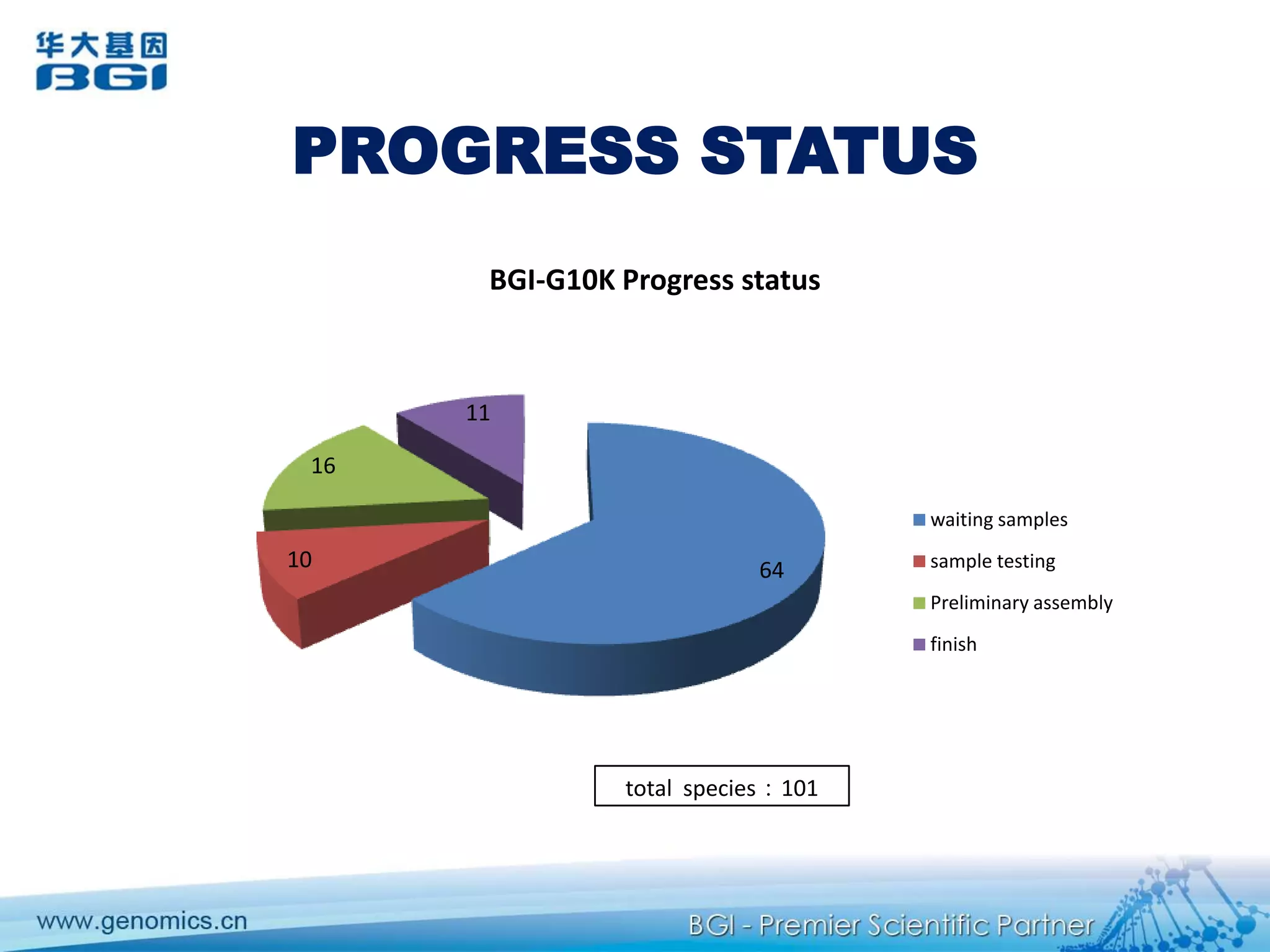
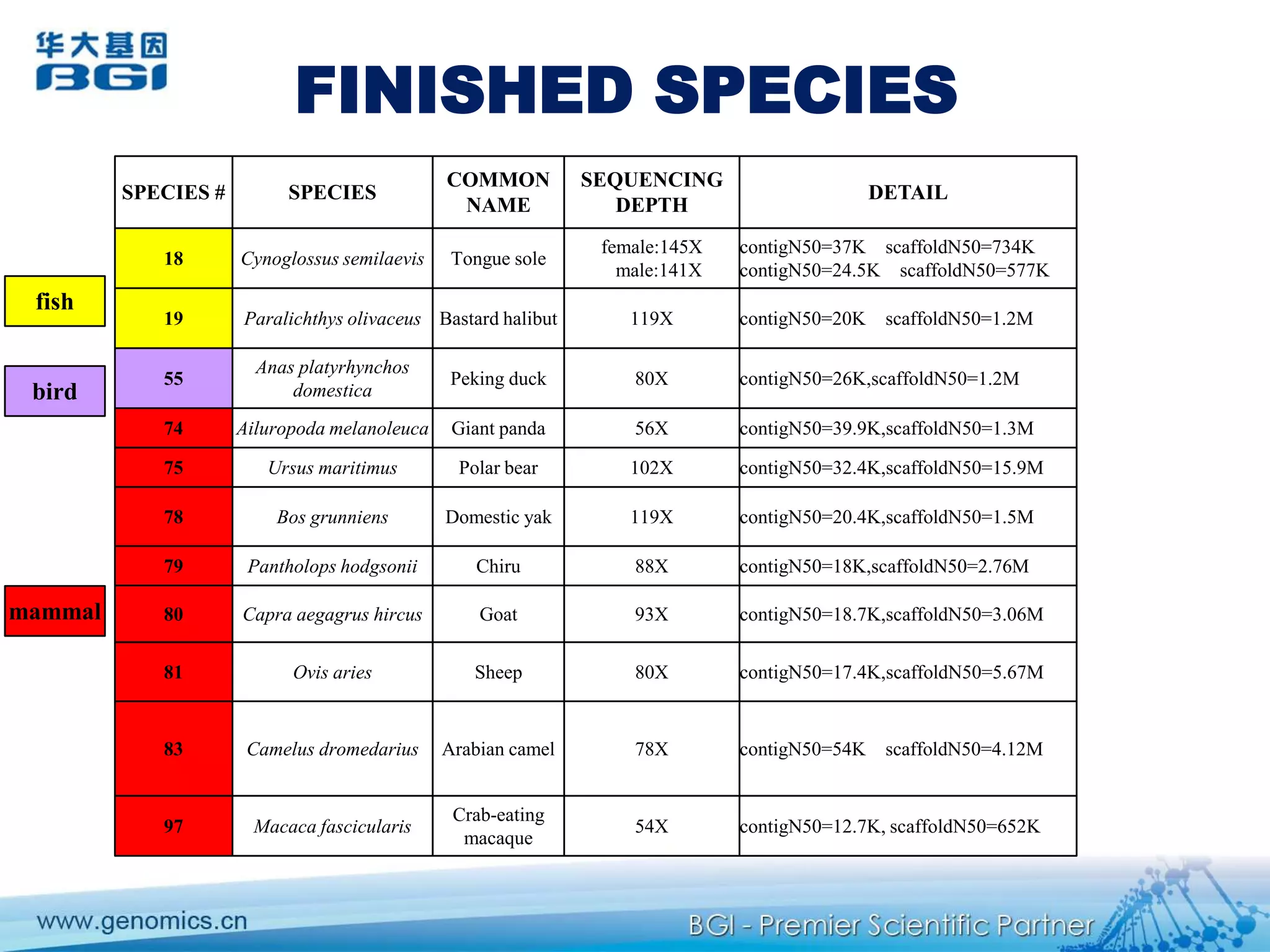
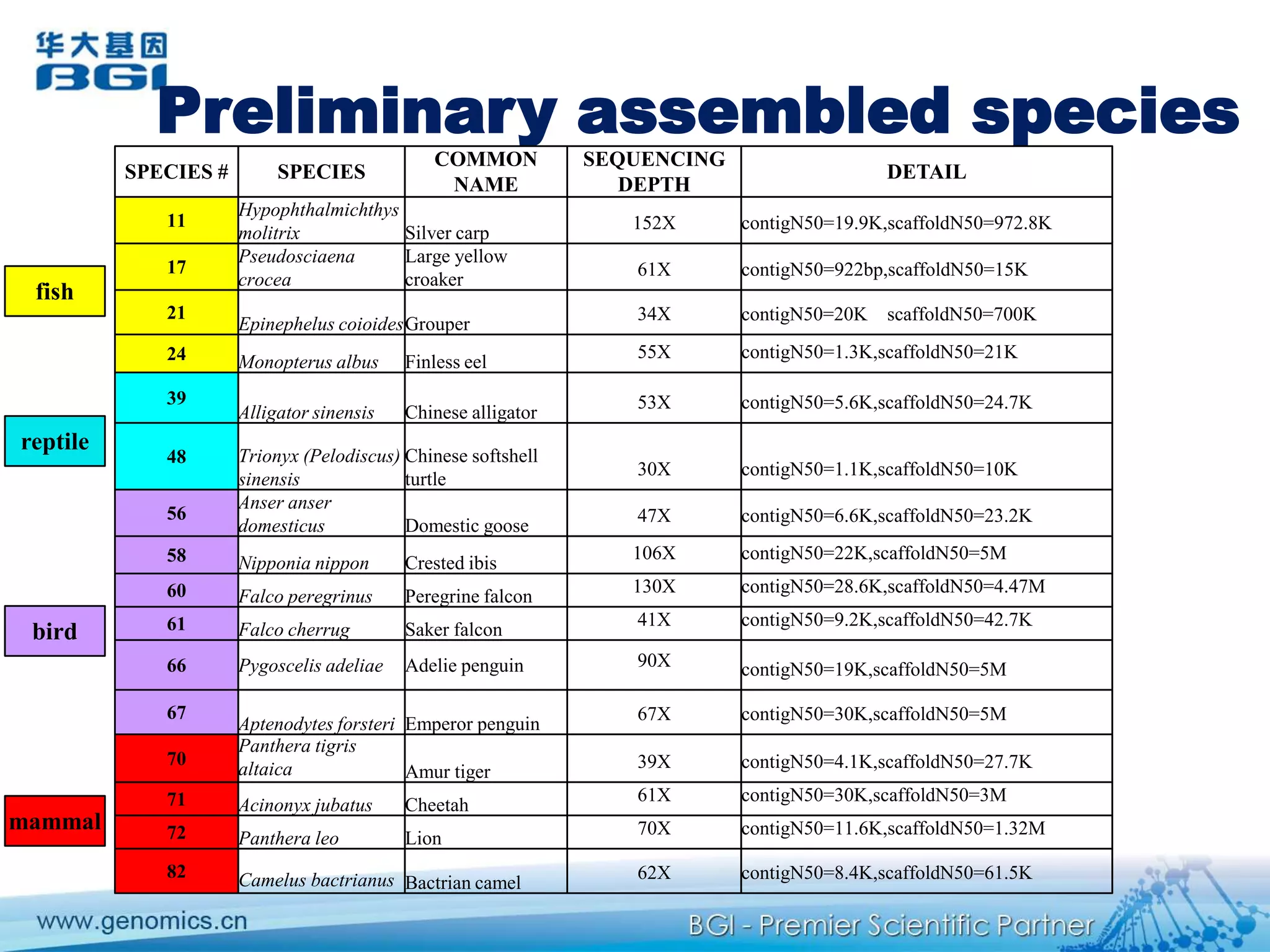
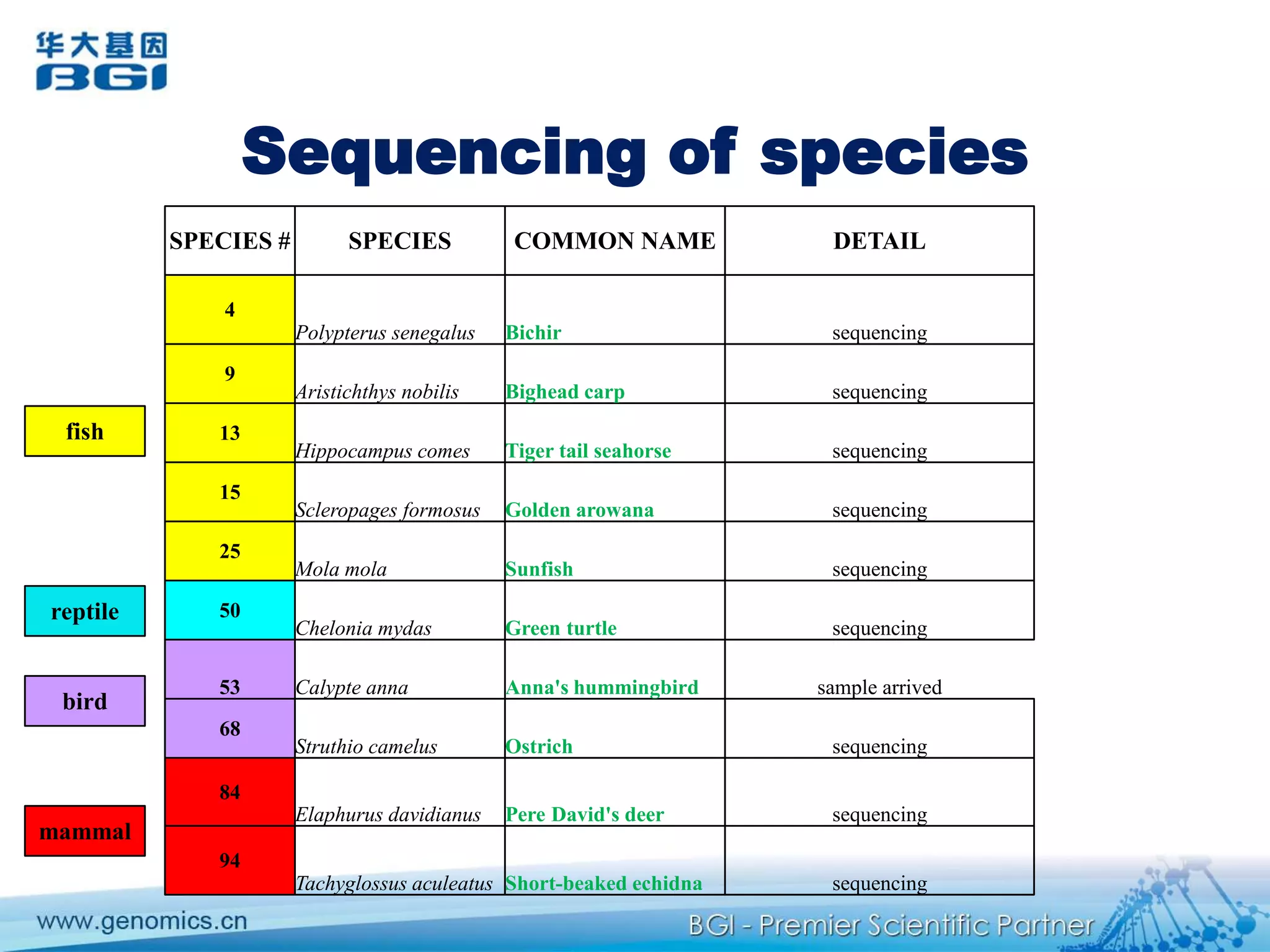
This document discusses the progresses and challenges of de novo genome assembly using next-generation sequencing data, including improvements made to error correction, contig construction, scaffolding, gap closure, and computational performance that have increased assembly quality and scalability; however, challenges still remain around resolving repeats and assembling heterozygous diploid genomes accurately.