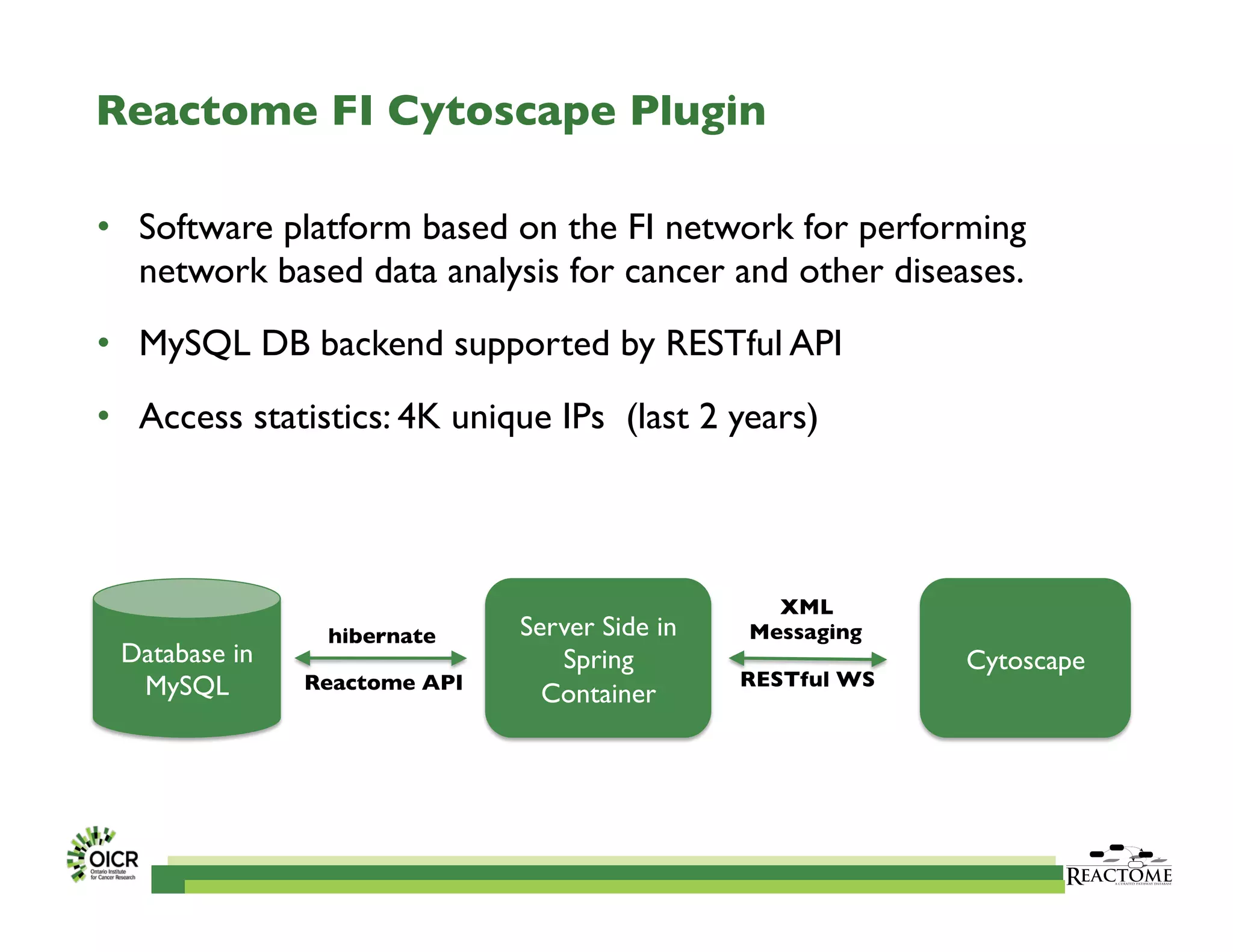
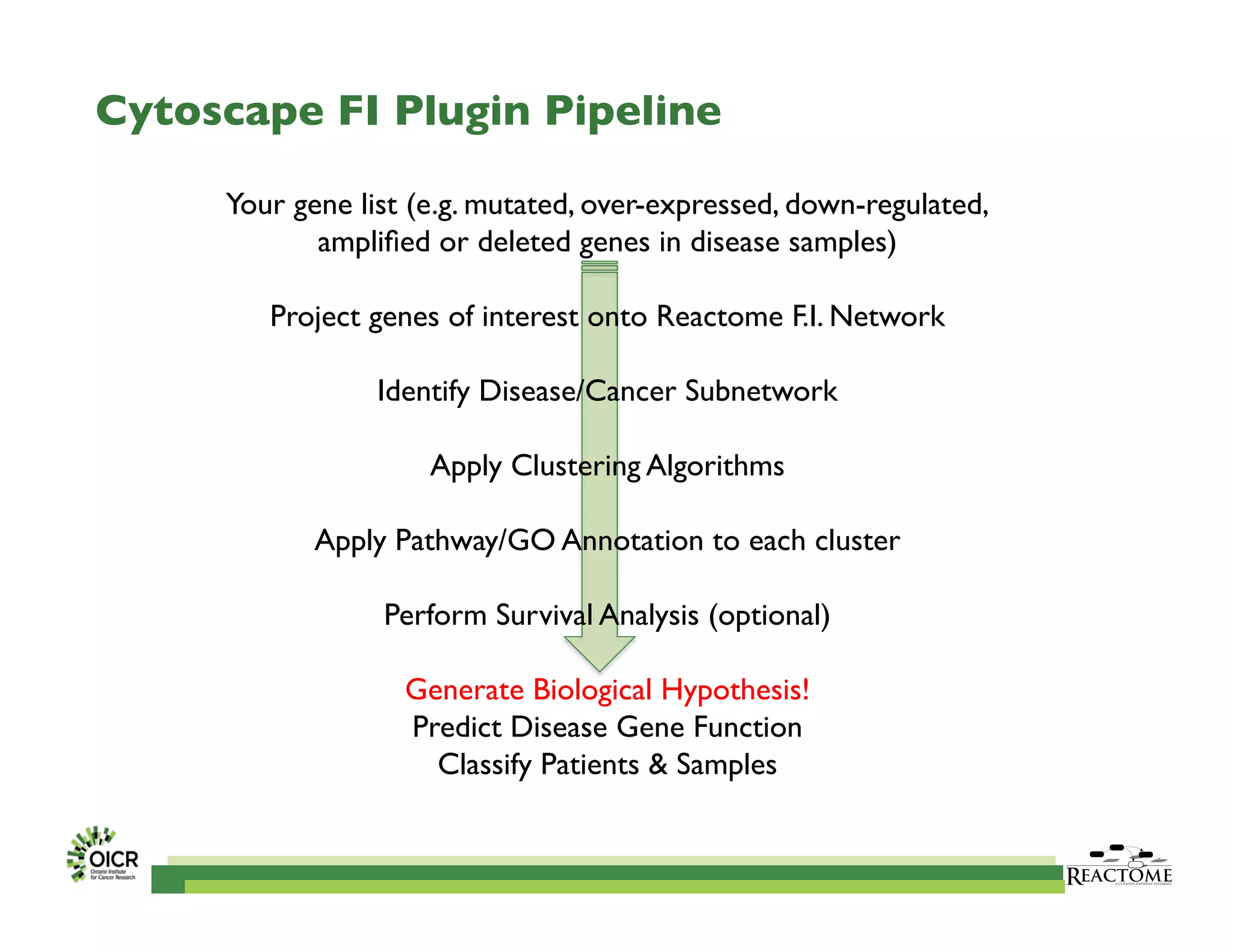
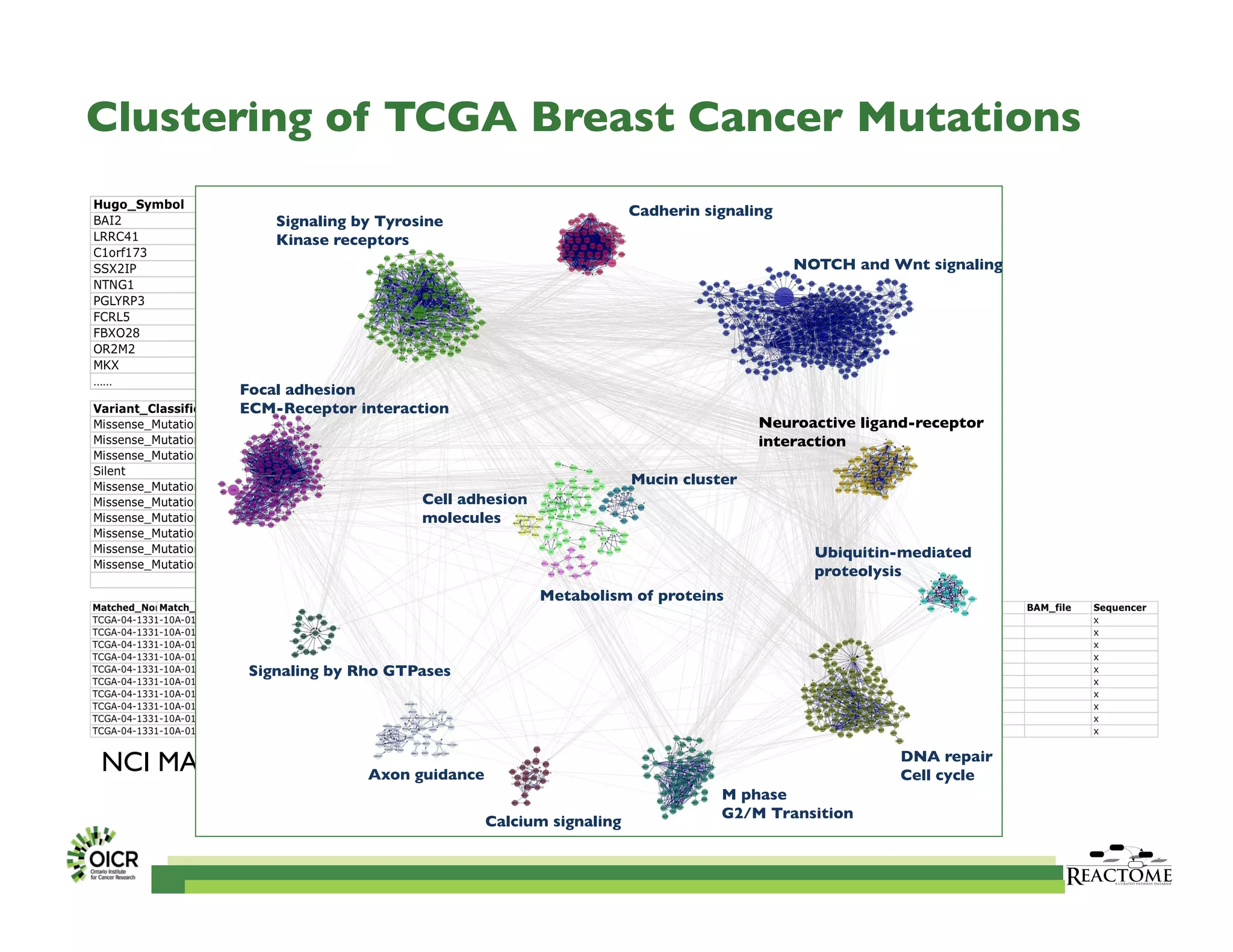
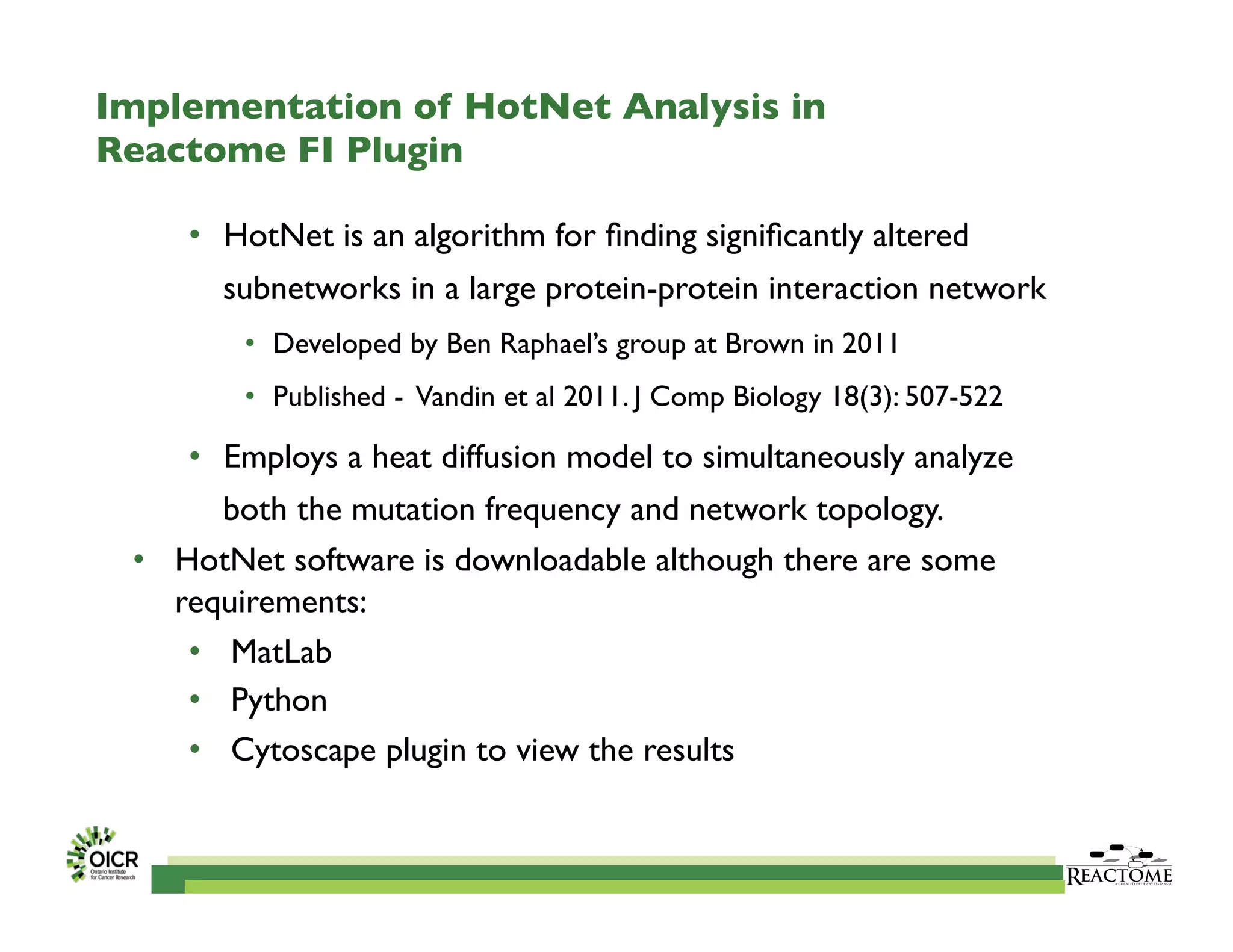
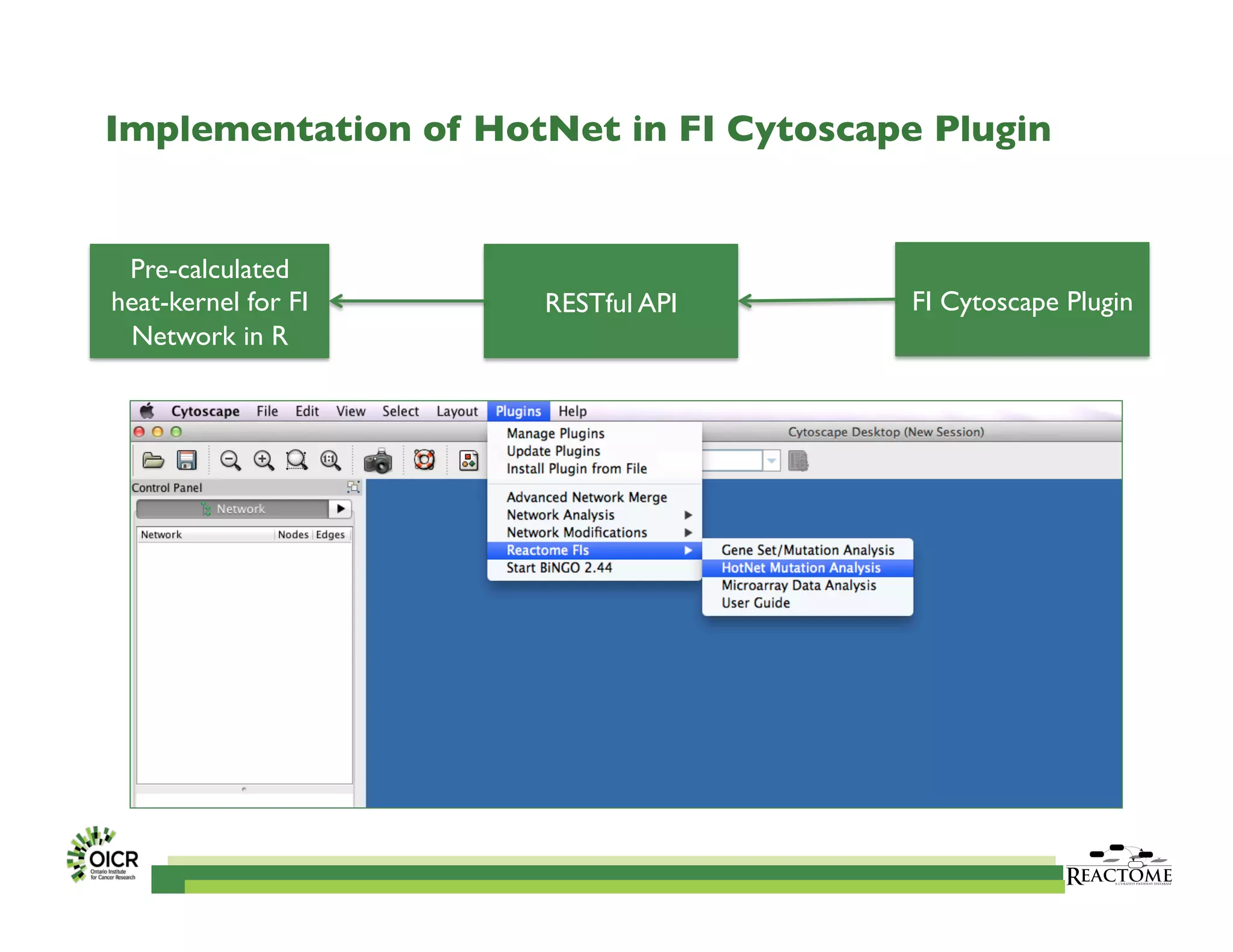
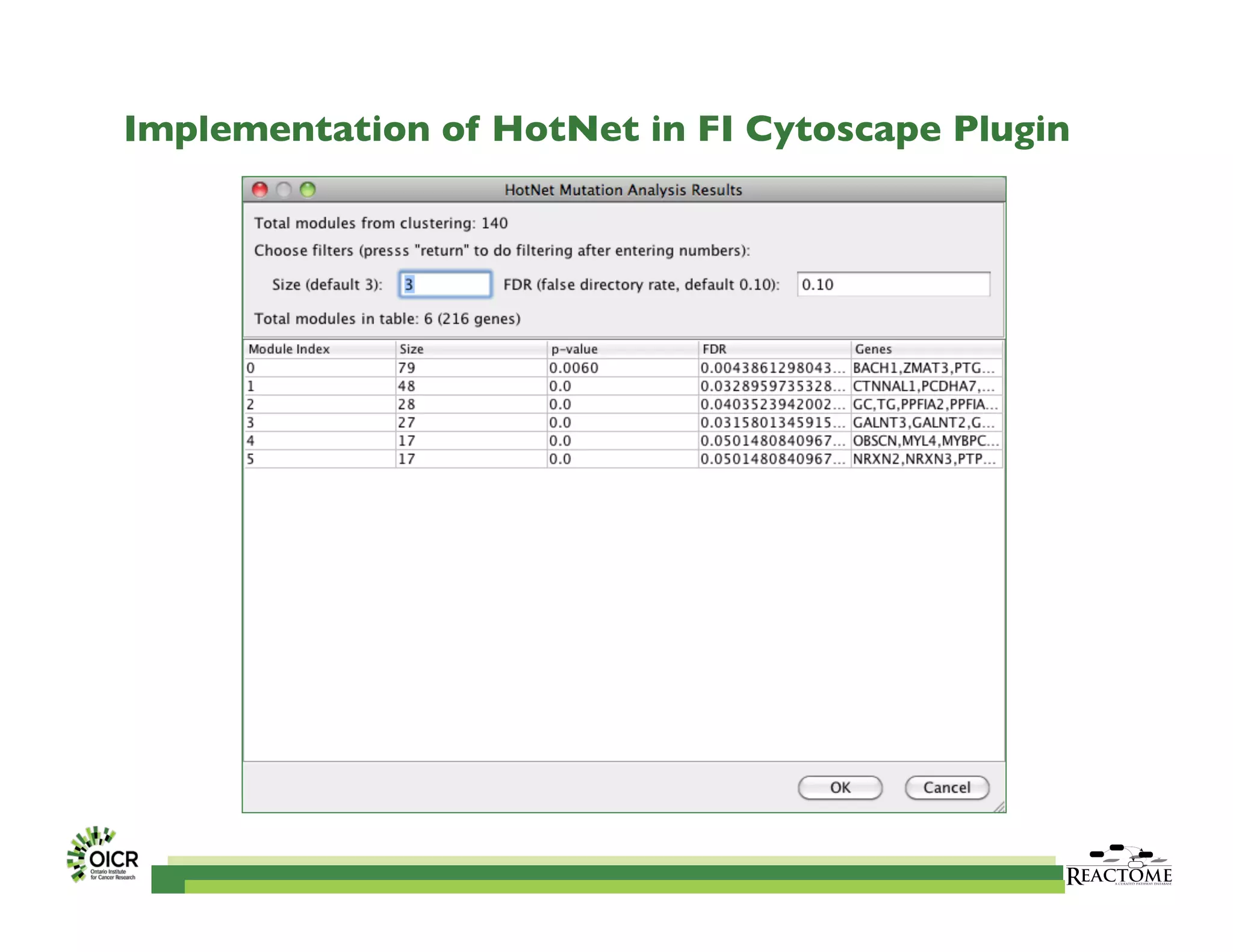
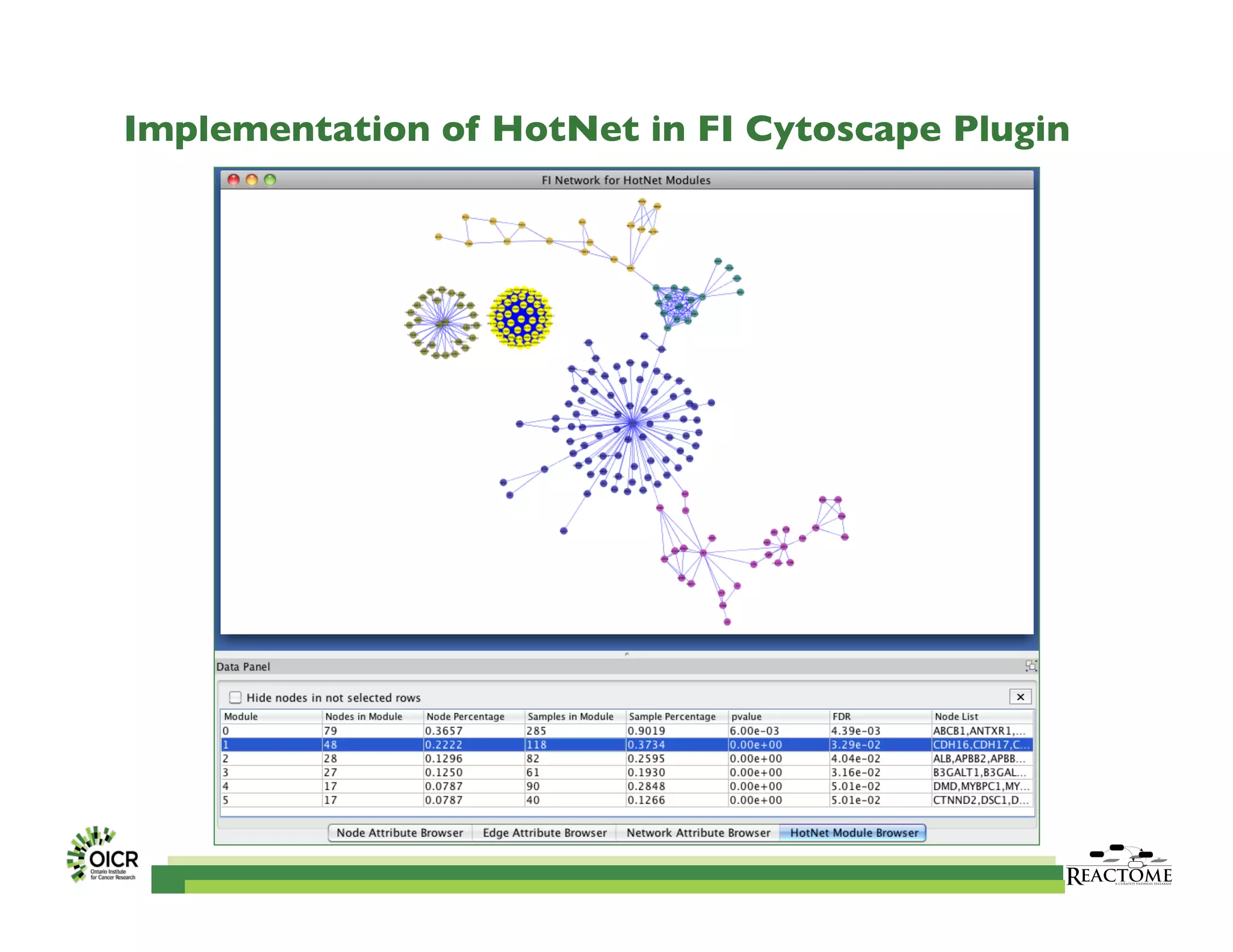
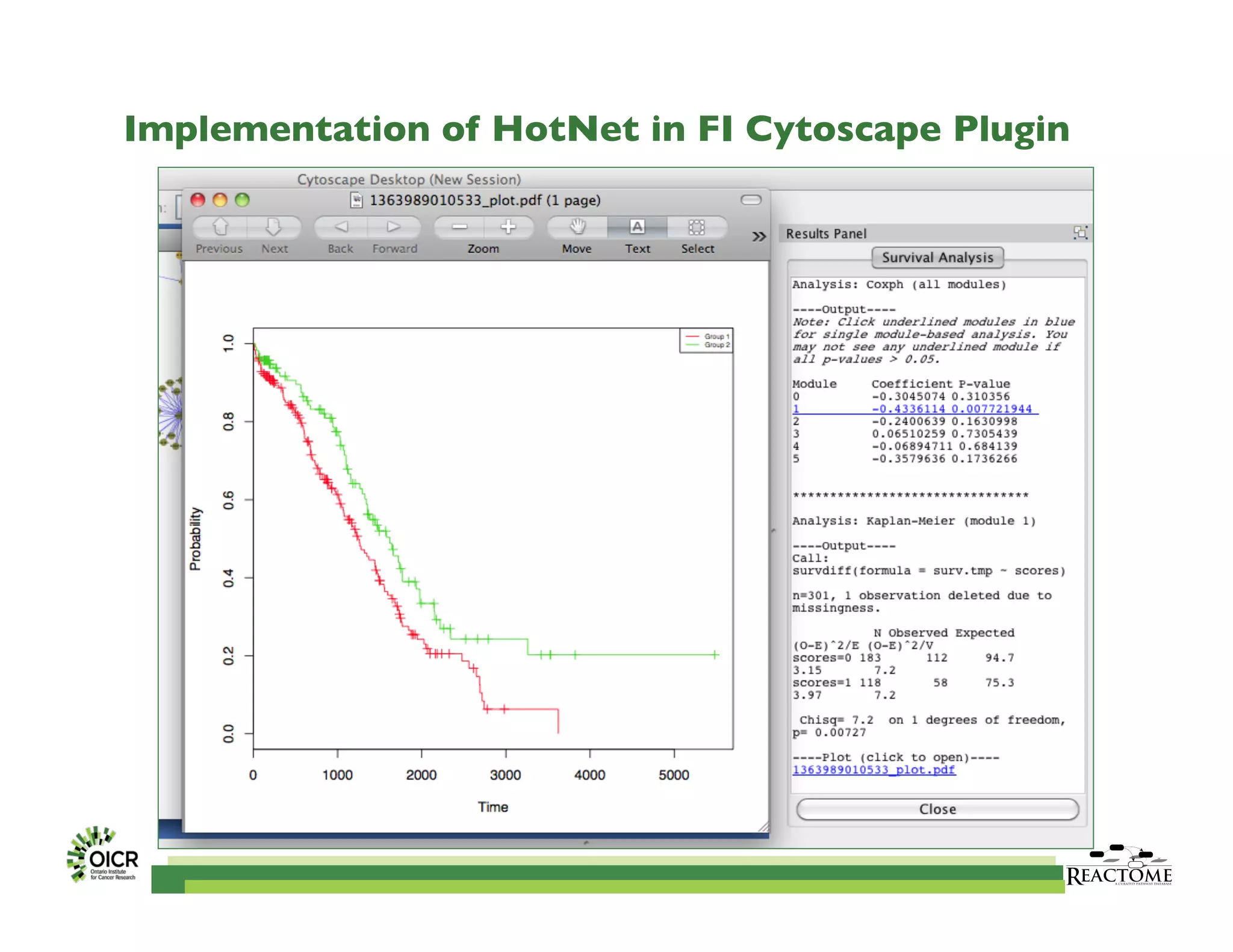
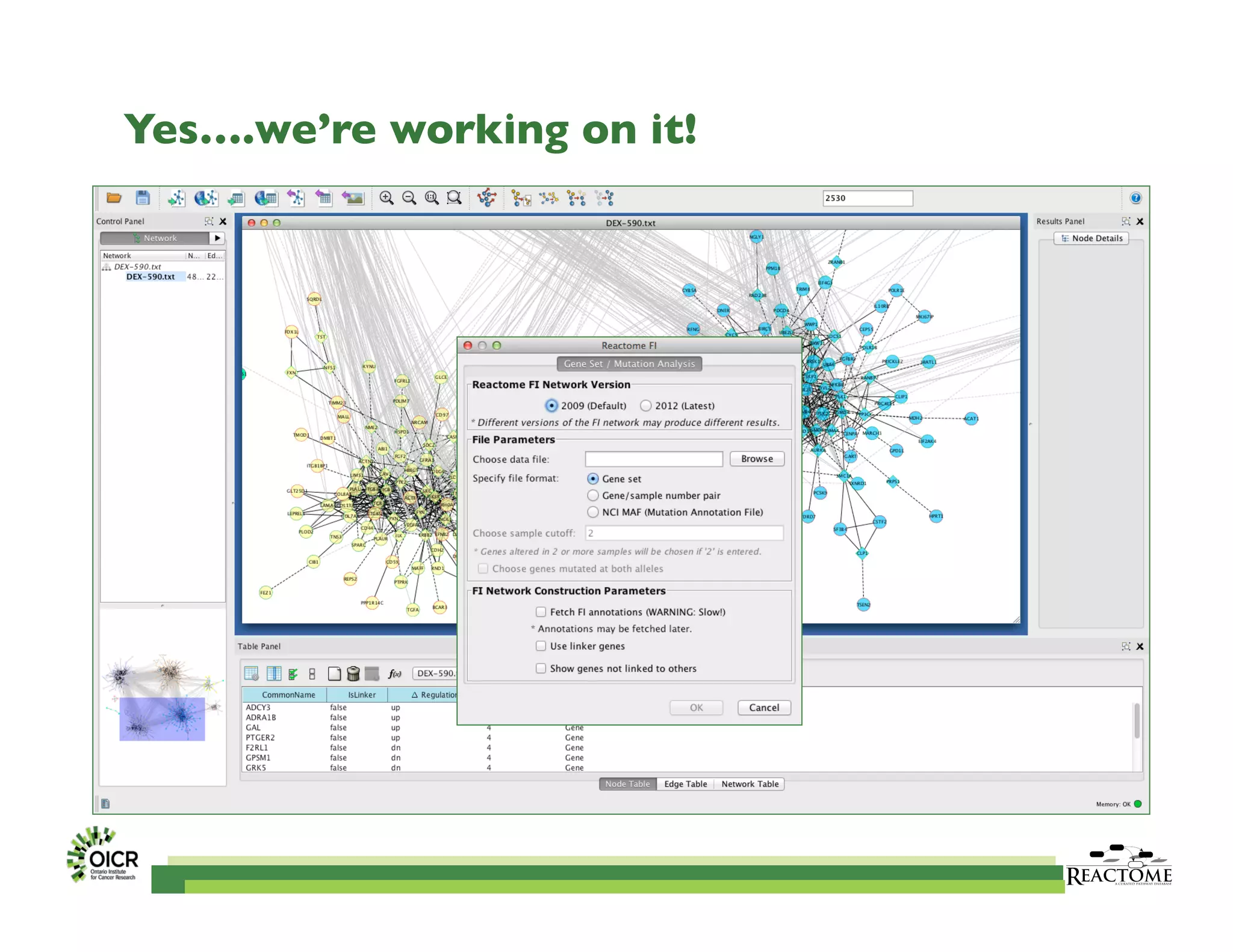
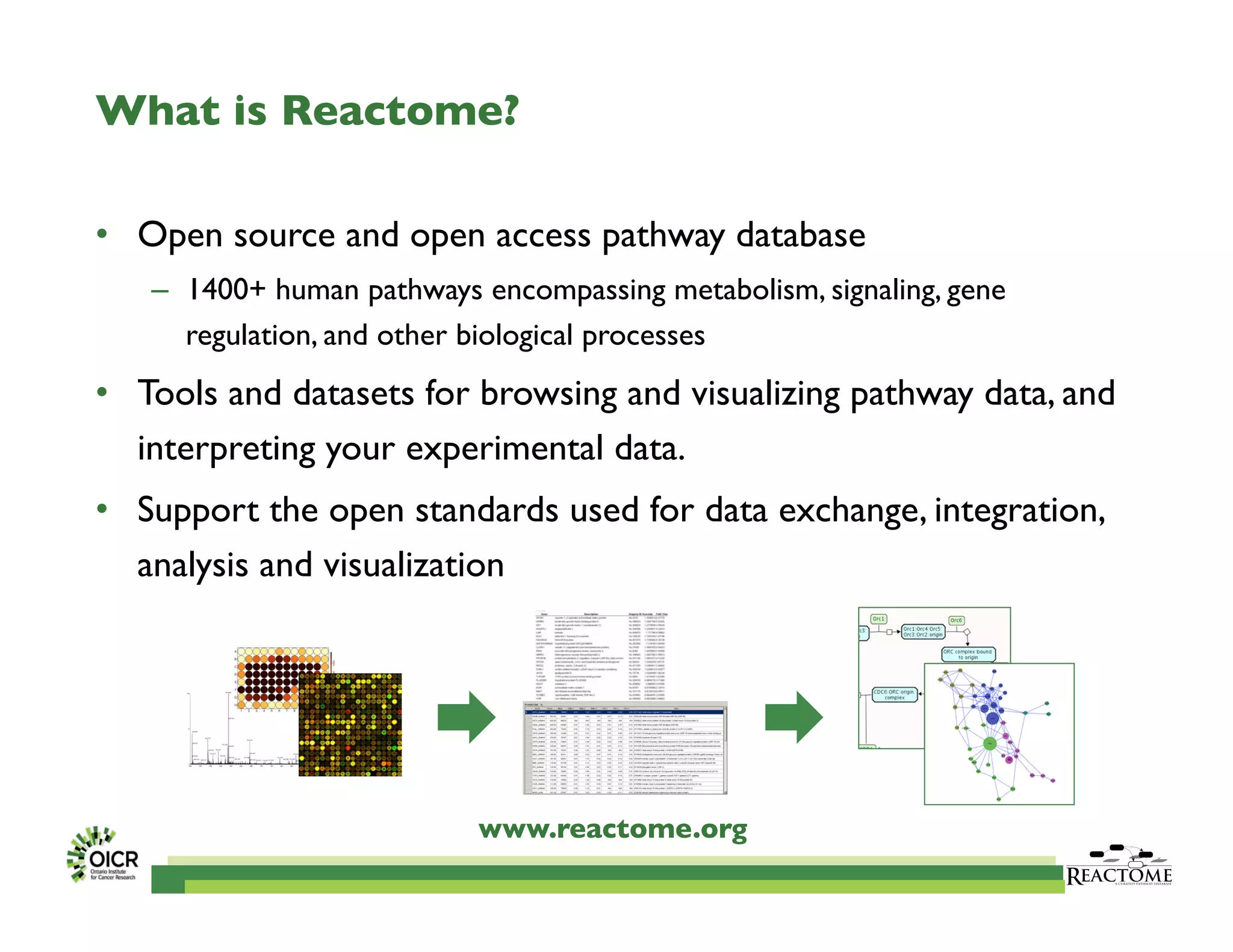
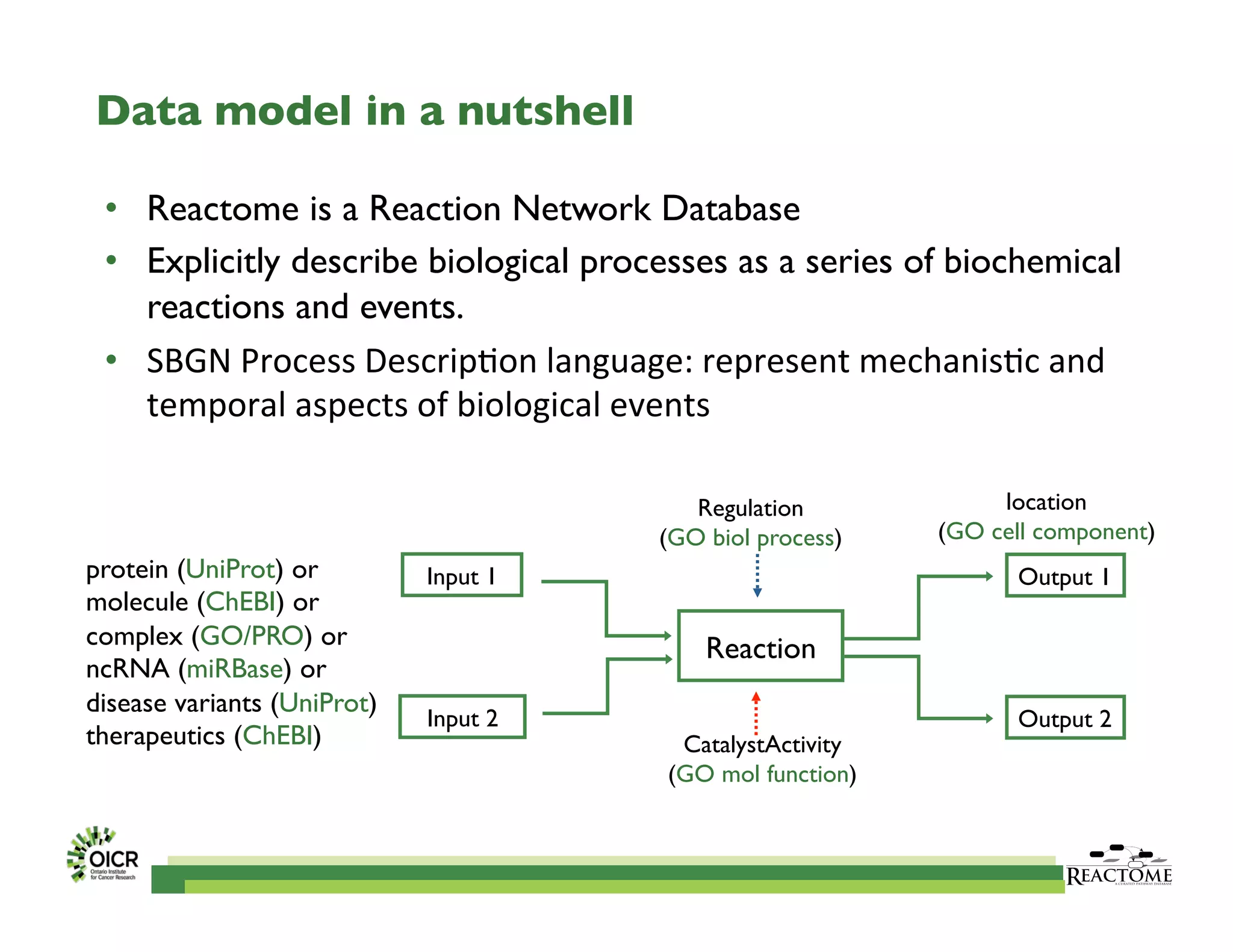
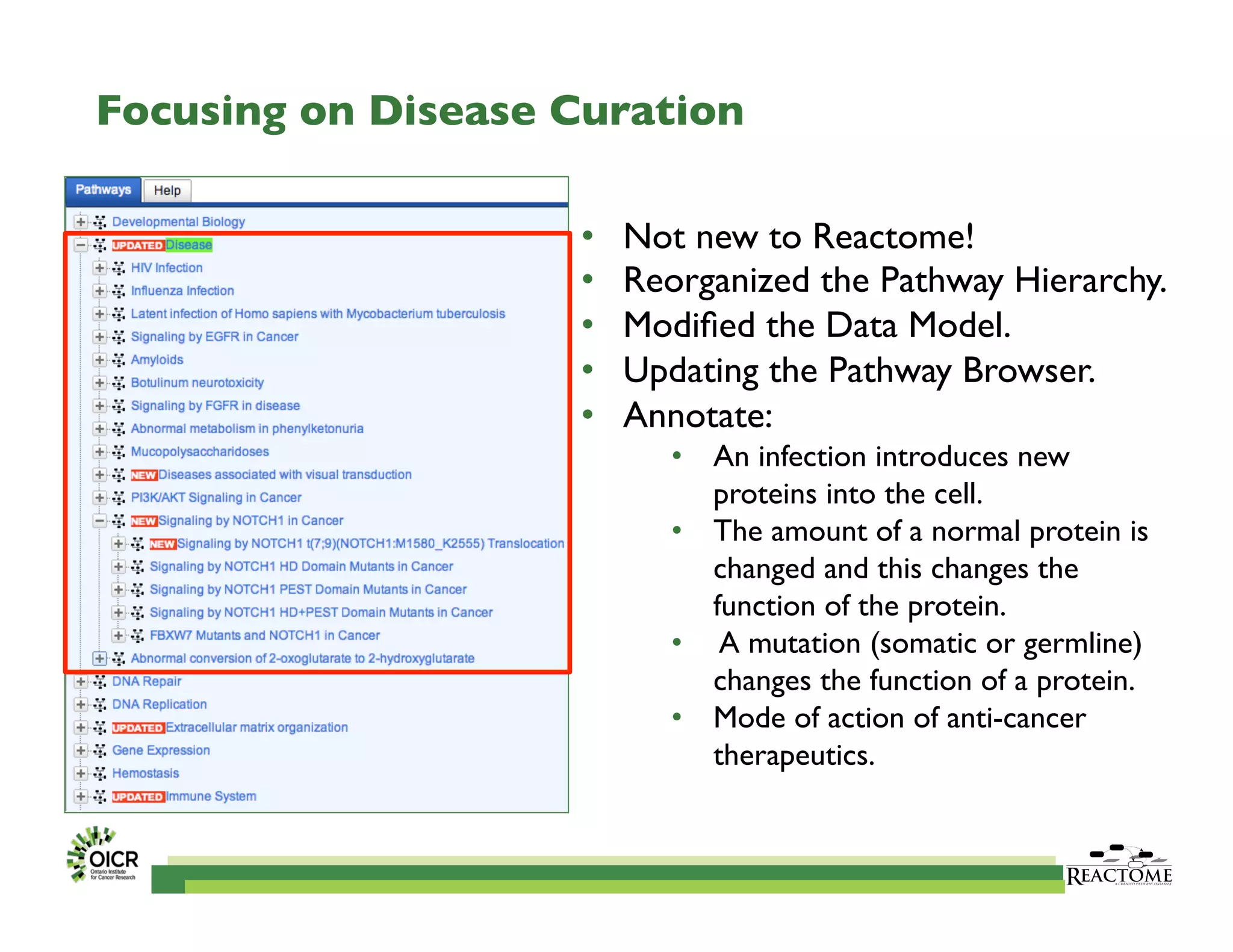
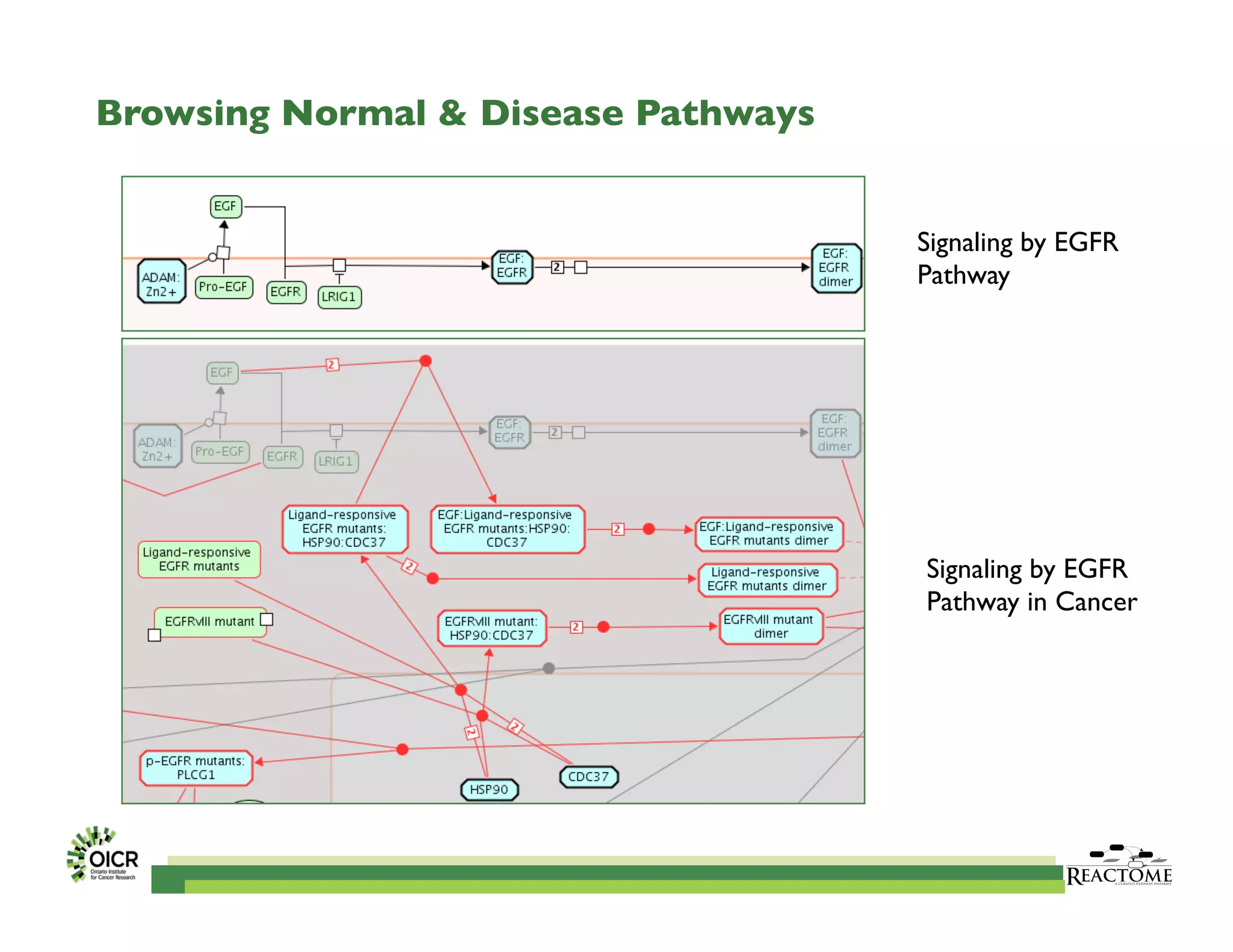
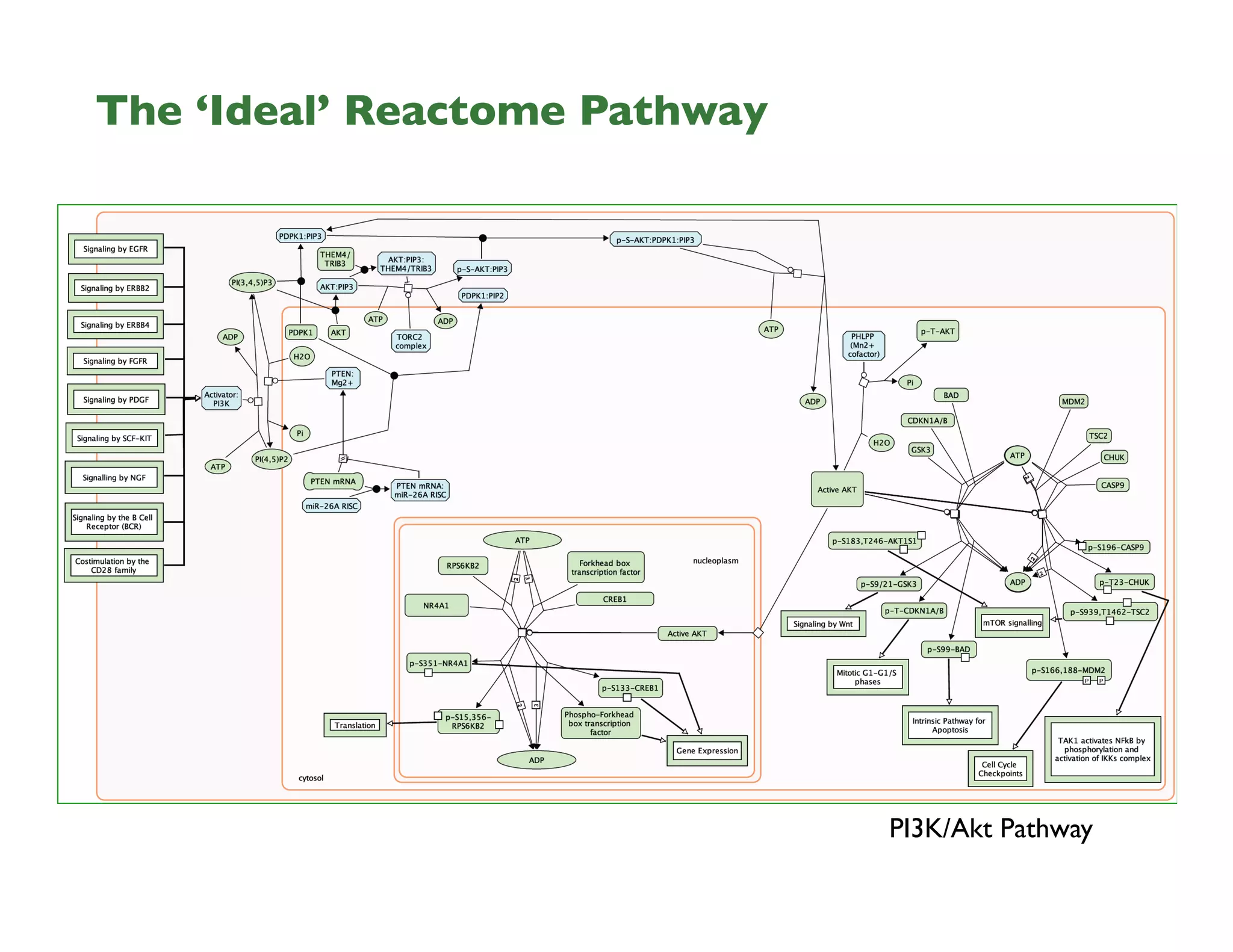
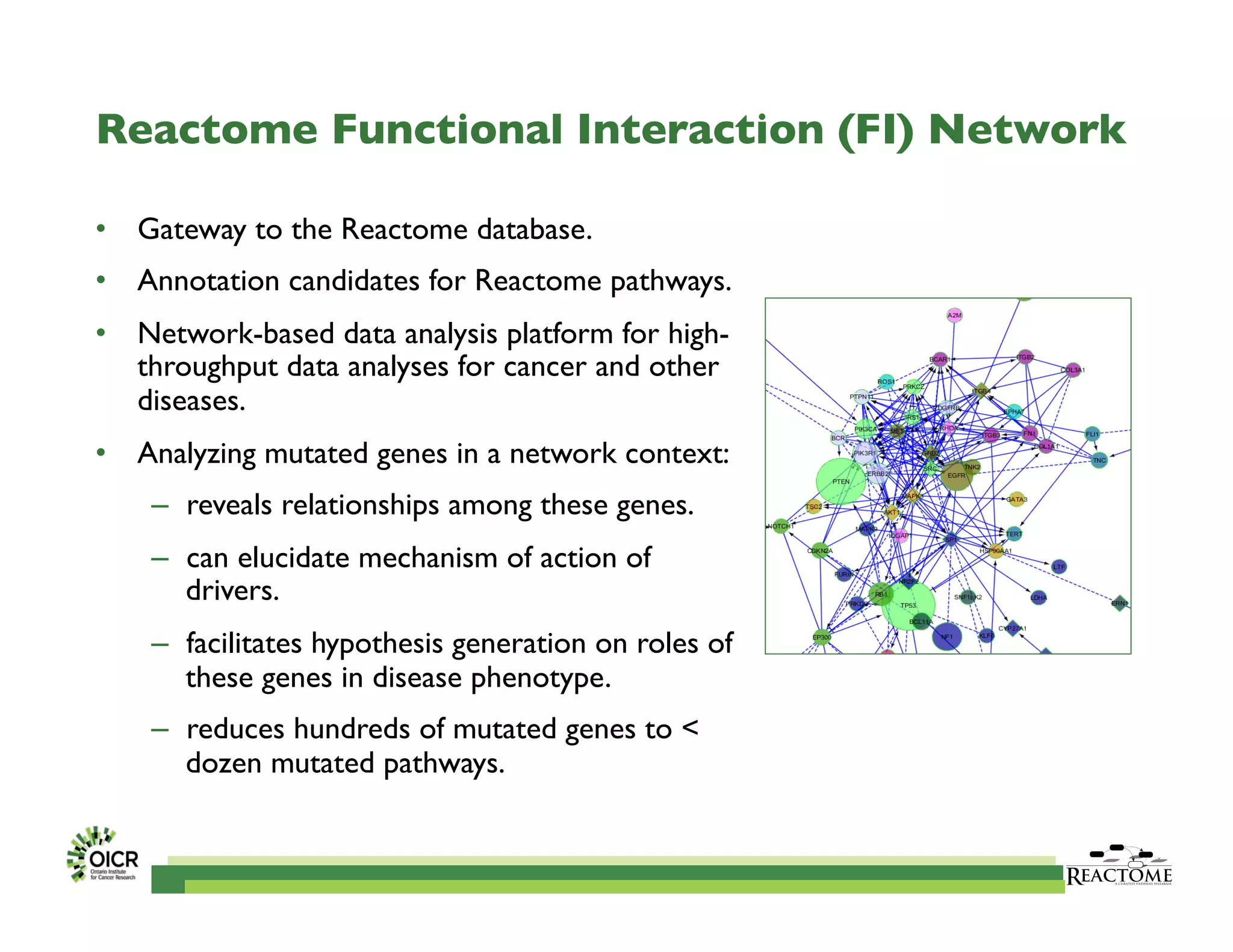
Reactome is an open-source pathway database that provides tools for visualizing and interpreting biological processes, including over 1400 human pathways related to metabolism and signaling. The Reactome Functional Interaction (FI) network facilitates high-throughput data analysis in disease contexts, enabling users to analyze genes and pathways effectively. The ongoing development aims to enhance data curation, integrate various data types, and improve user experience on the platform.
![Creation of the Reactome FI Network
Human PPI [45-47] Fly PPI [45]
Domain Interaction [52]
Prieto’s Gene Expression [50]Lee’s Gene Expression [49]
GO BP Sharing [51]Yeast PPI [45]
Worm PPI [45]
PPIs from GeneWays [53]
Data sources for Predicted FIs
Reactome [23]
Panther [60]
KEGG [63]
TRED [64]
NCI-BioCarta [62]
NCI-Nature [62]
CellMap [61]
Data sources for
Annotated FIs
Naïve Bayes
Classifier
trained by
validated by
Predicted FIs Annotated FIs
Reactome FI
Network
Mouse
PPI
2,3
2
2
2,3
2
2,3
ENCODE
TF/Target
273K interactions
and 11K proteins](https://image.slidesharecdn.com/2013reactomenetbiosig-130730201208-phpapp02/75/NetBioSIG2013-Talk-Robin-Haw-9-2048.jpg)