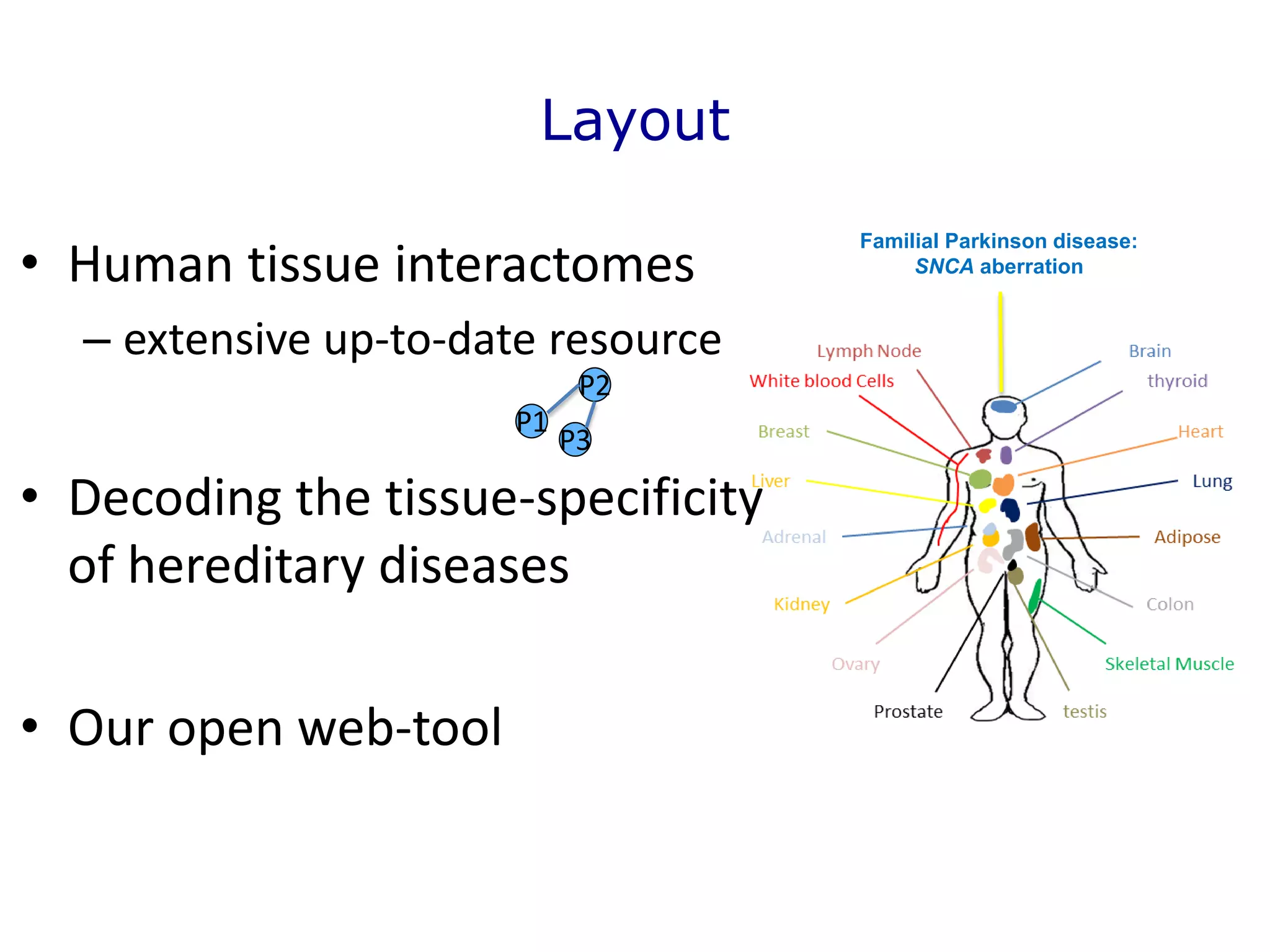
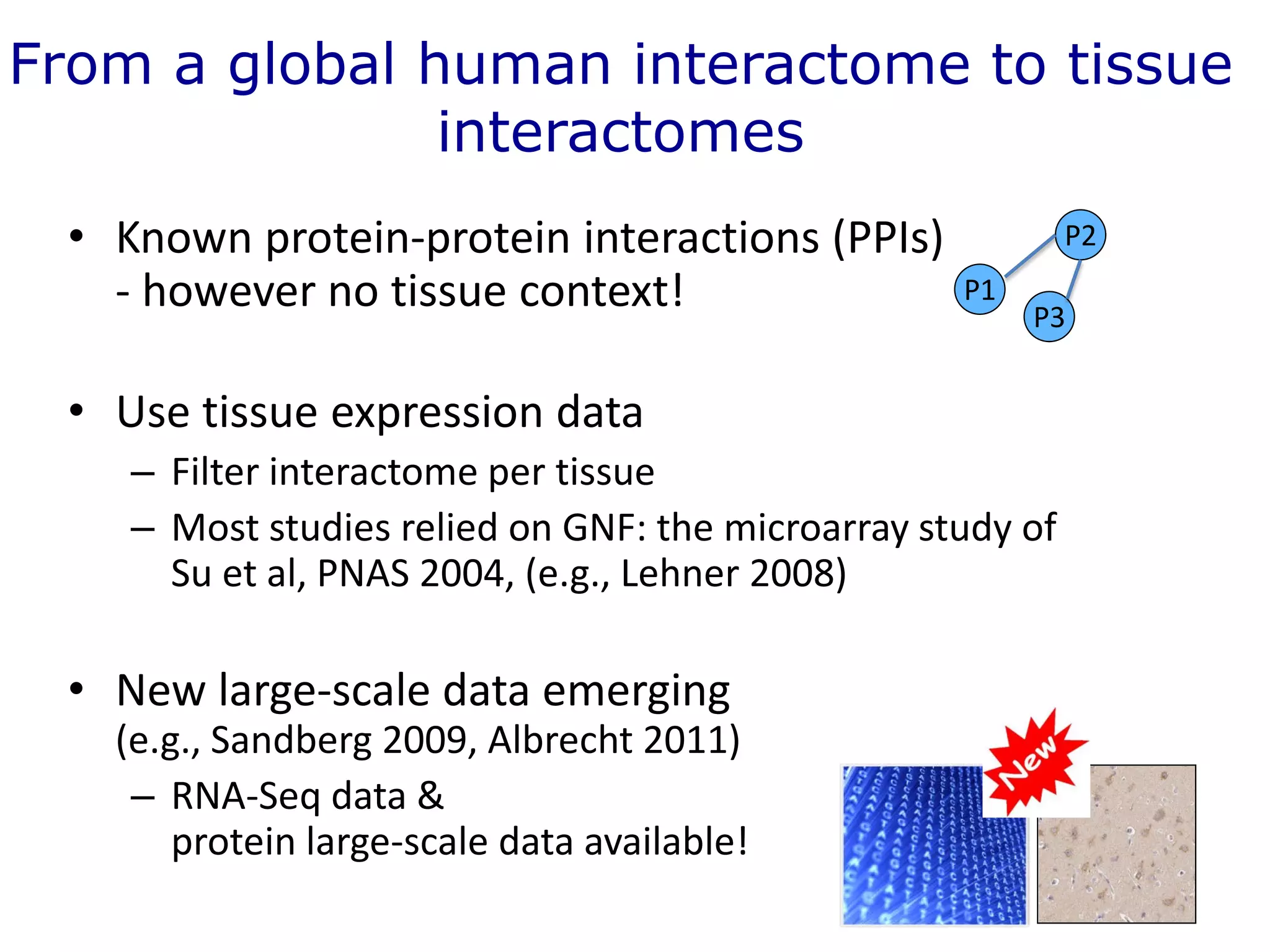
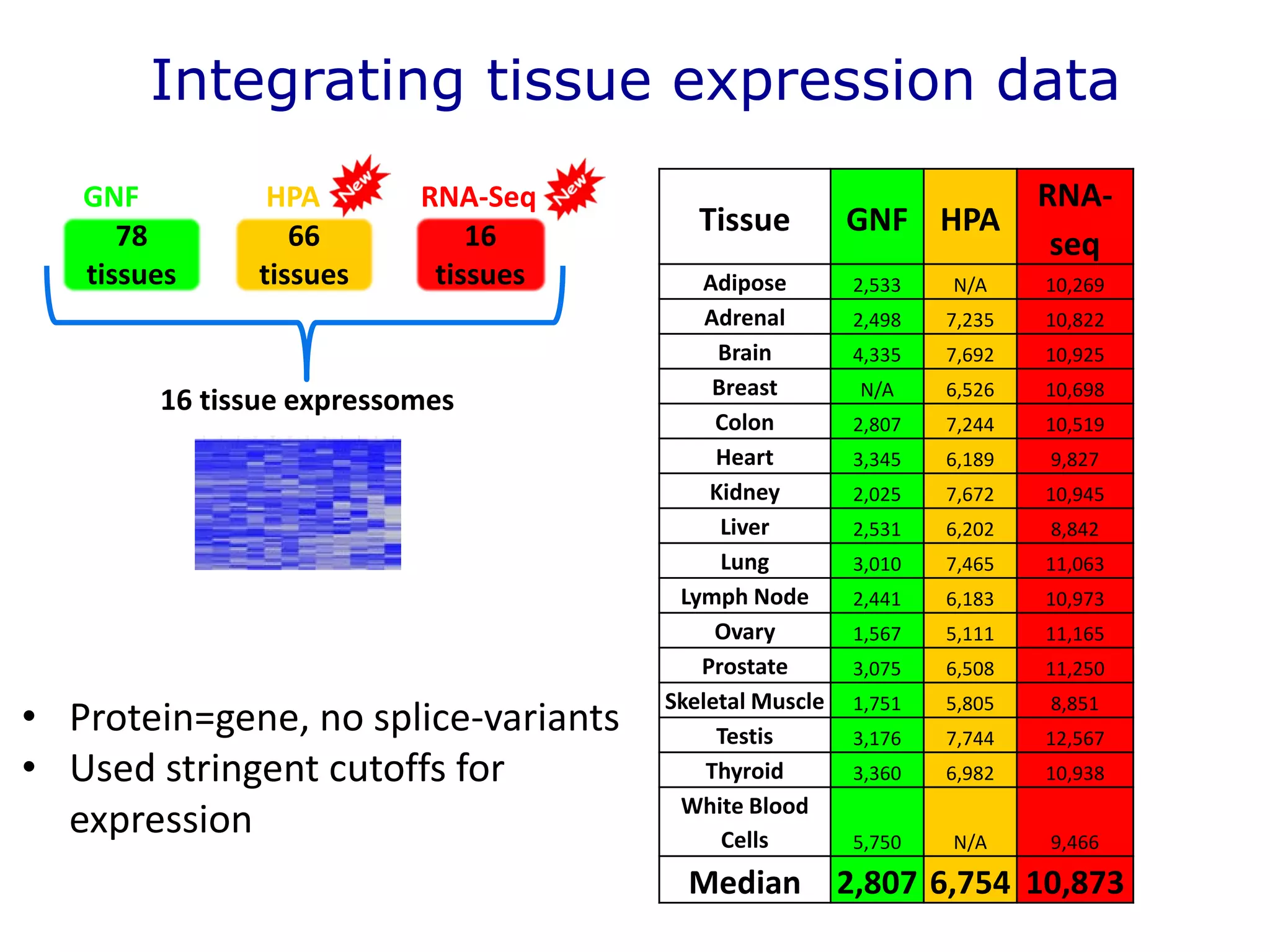
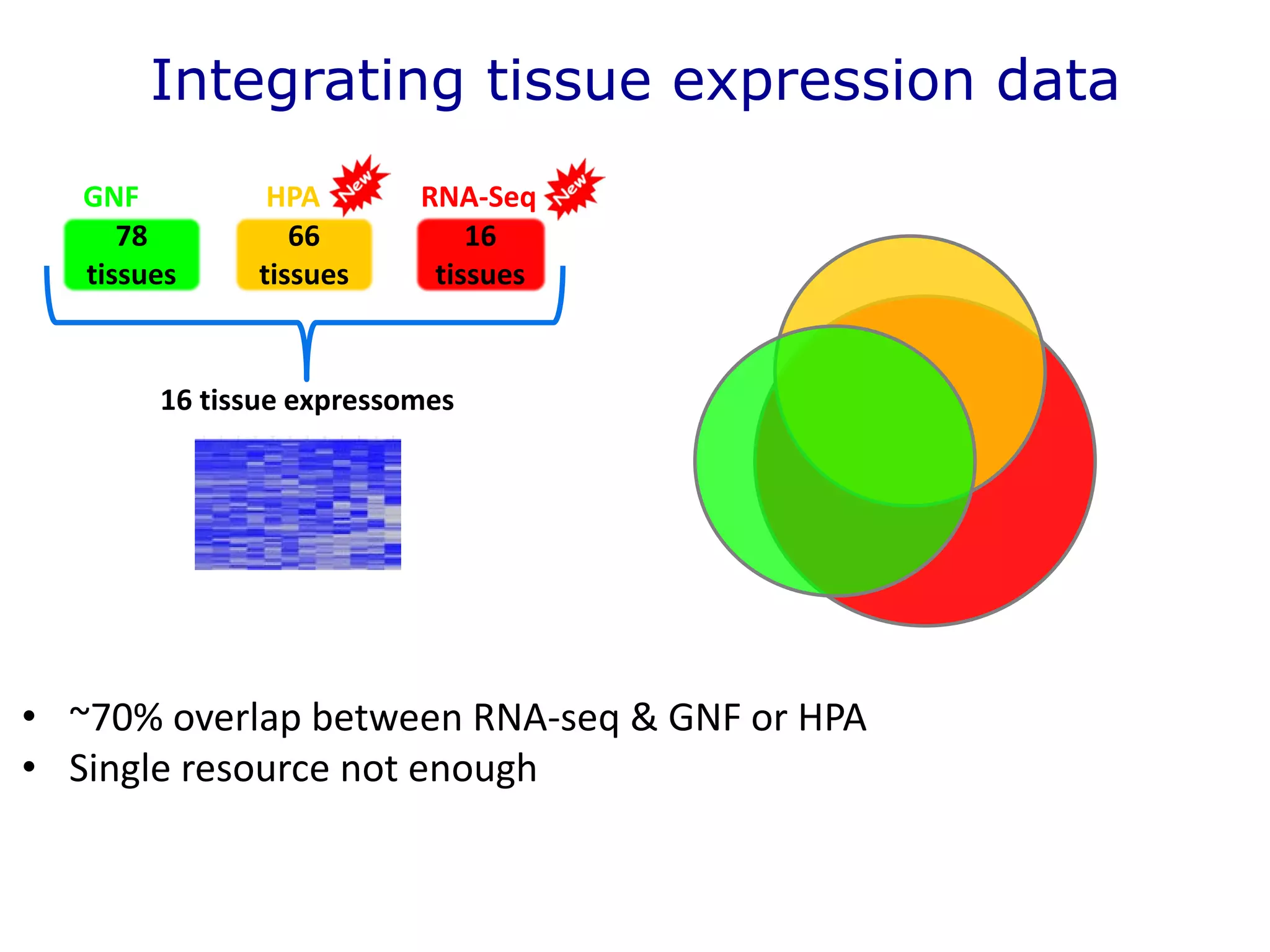
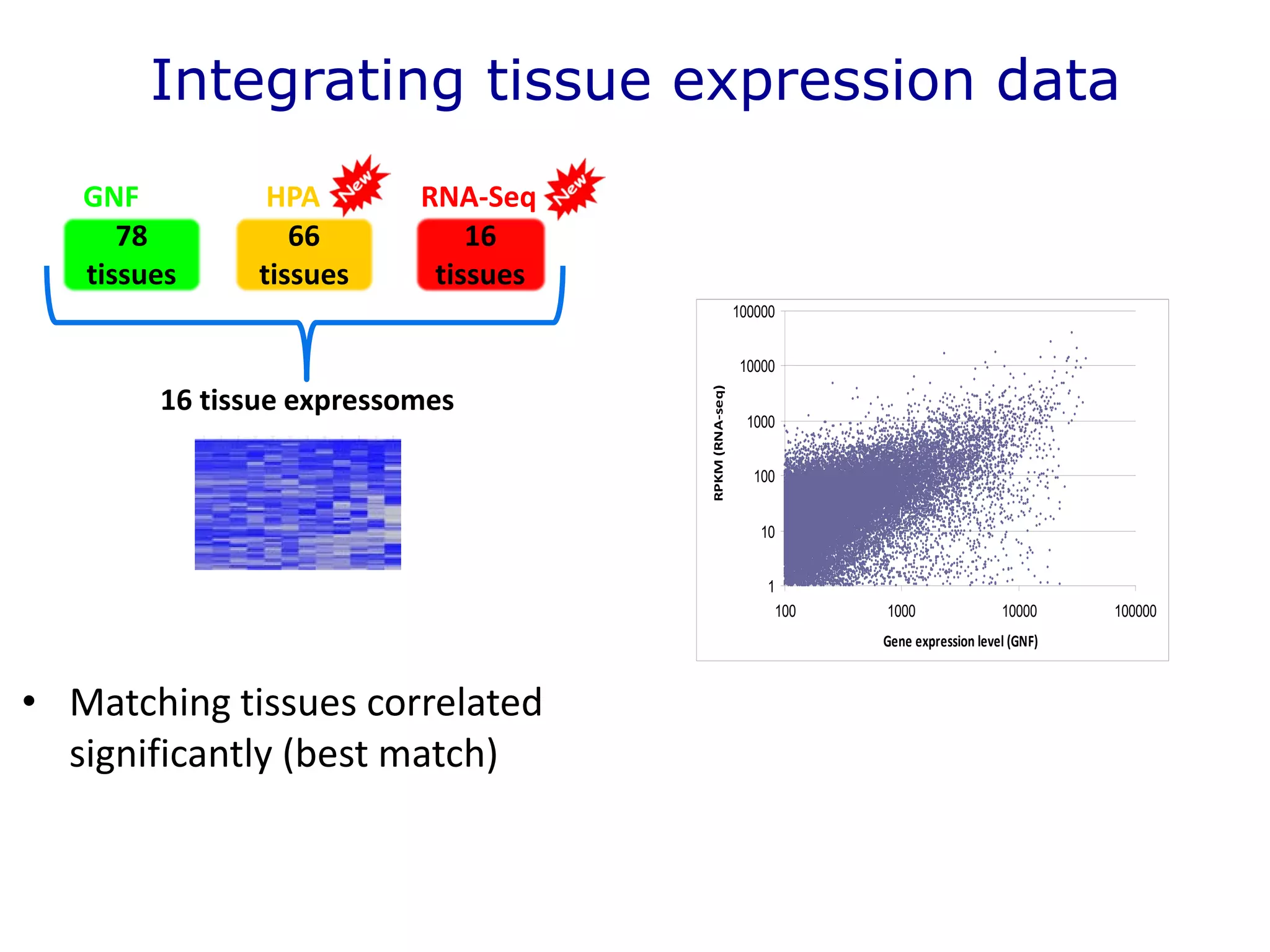
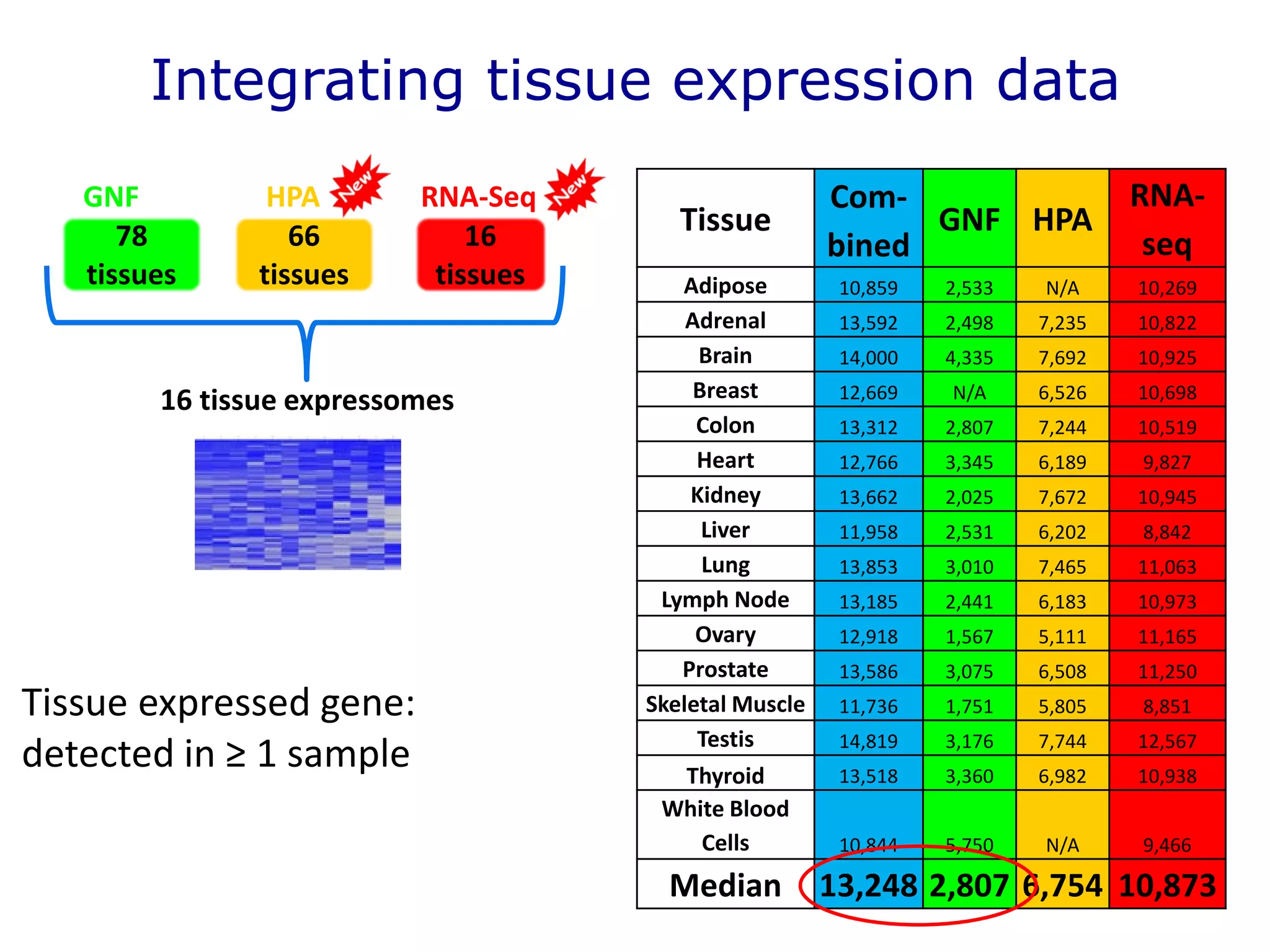
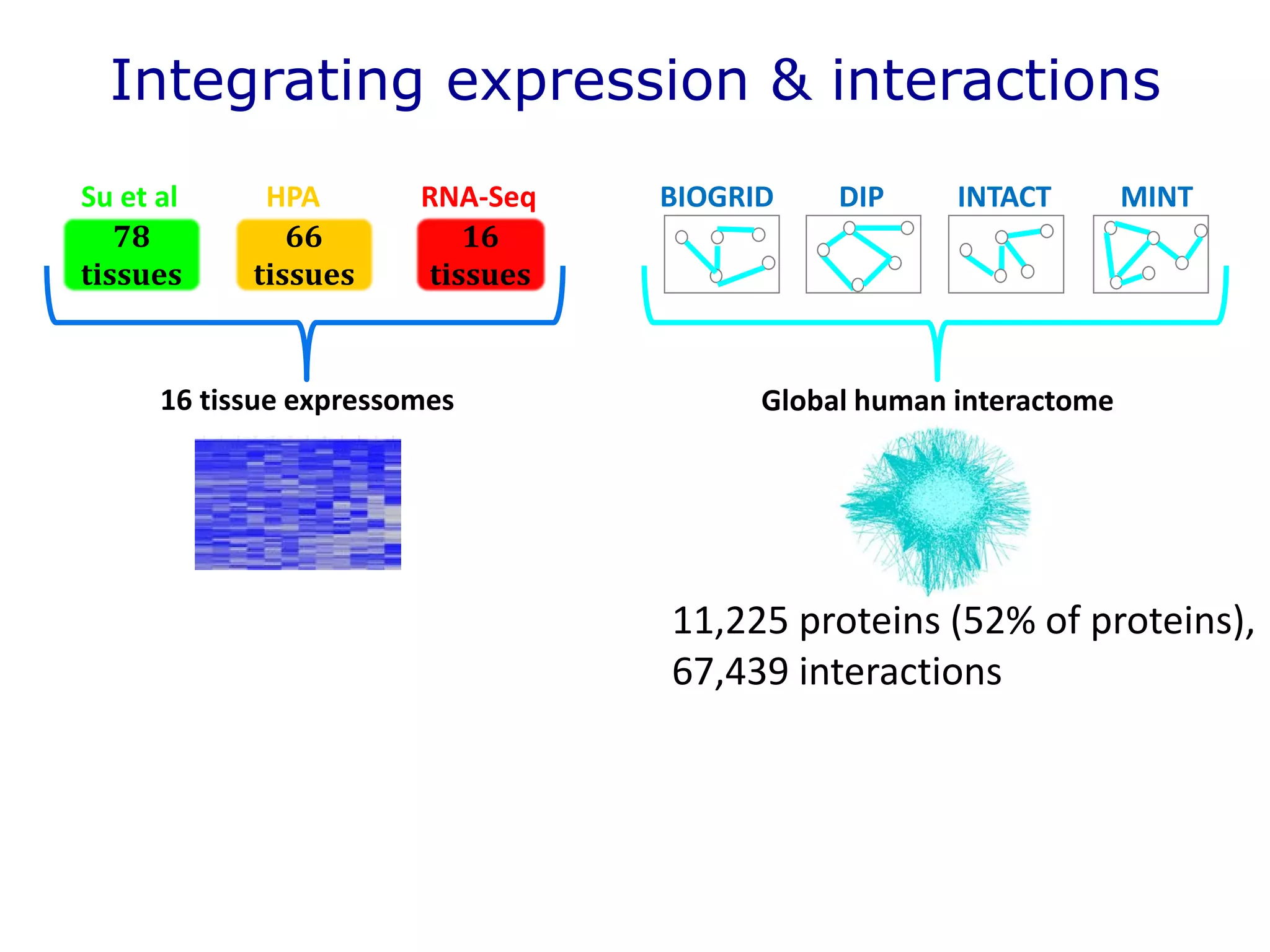
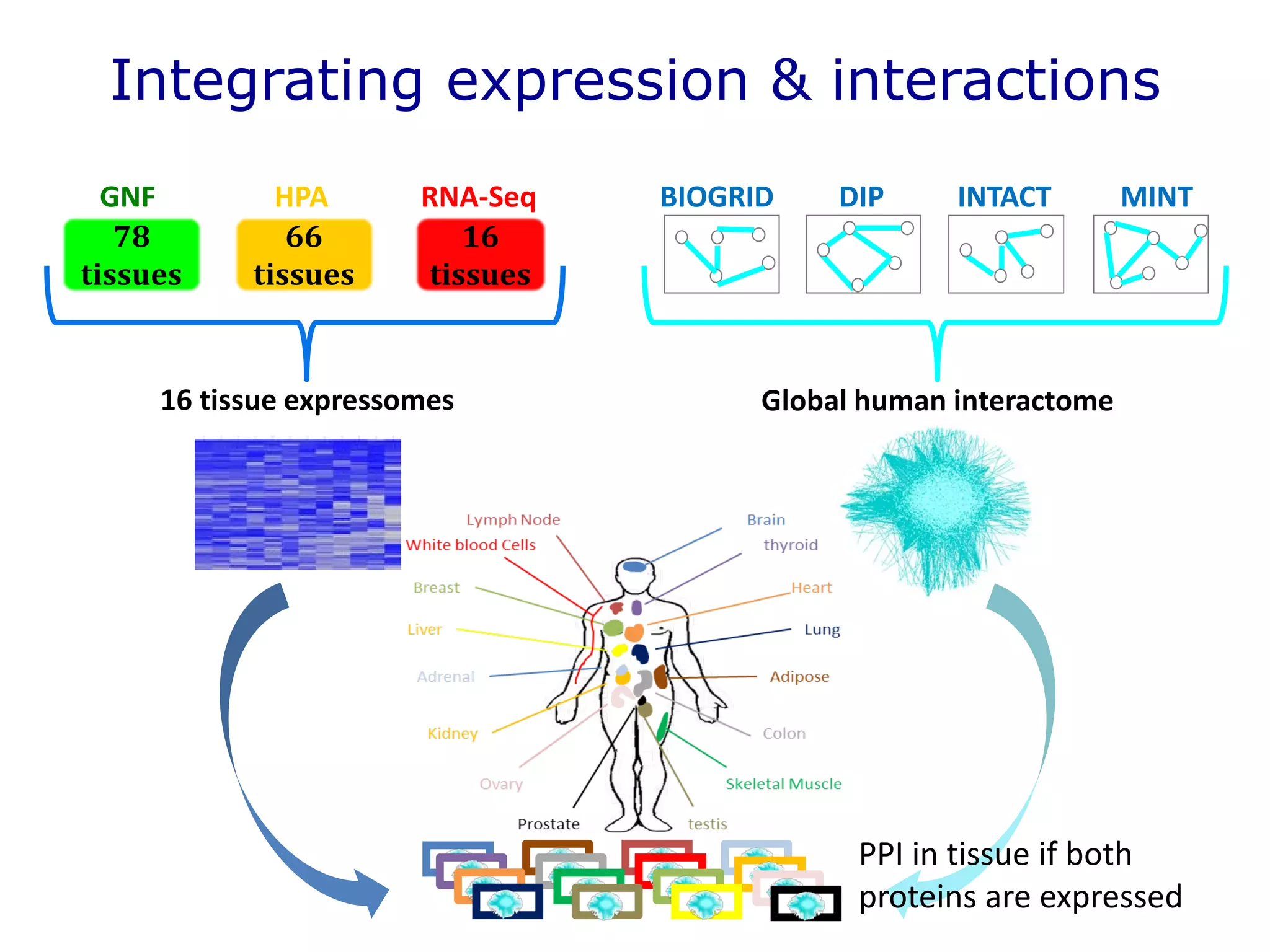
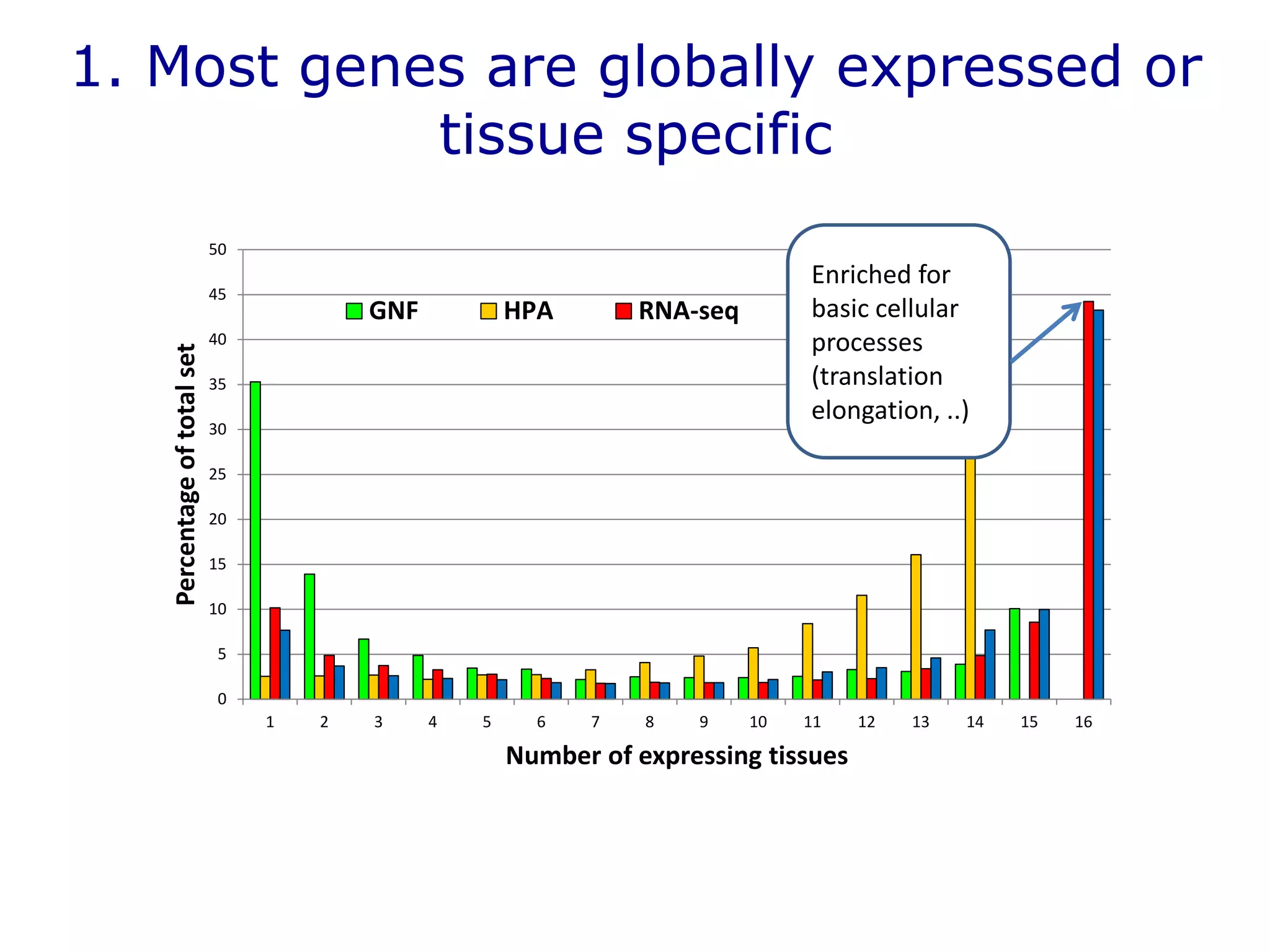
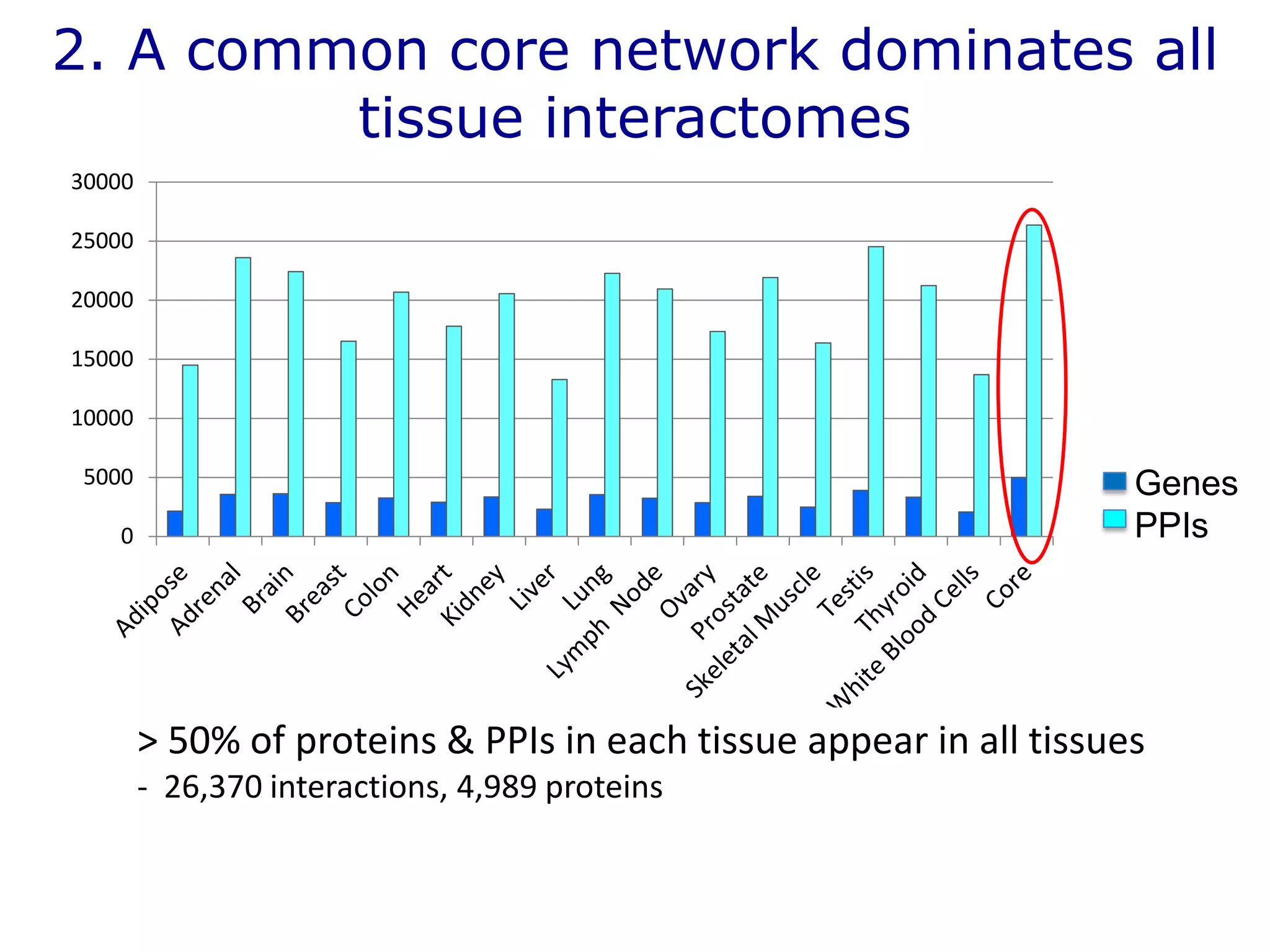
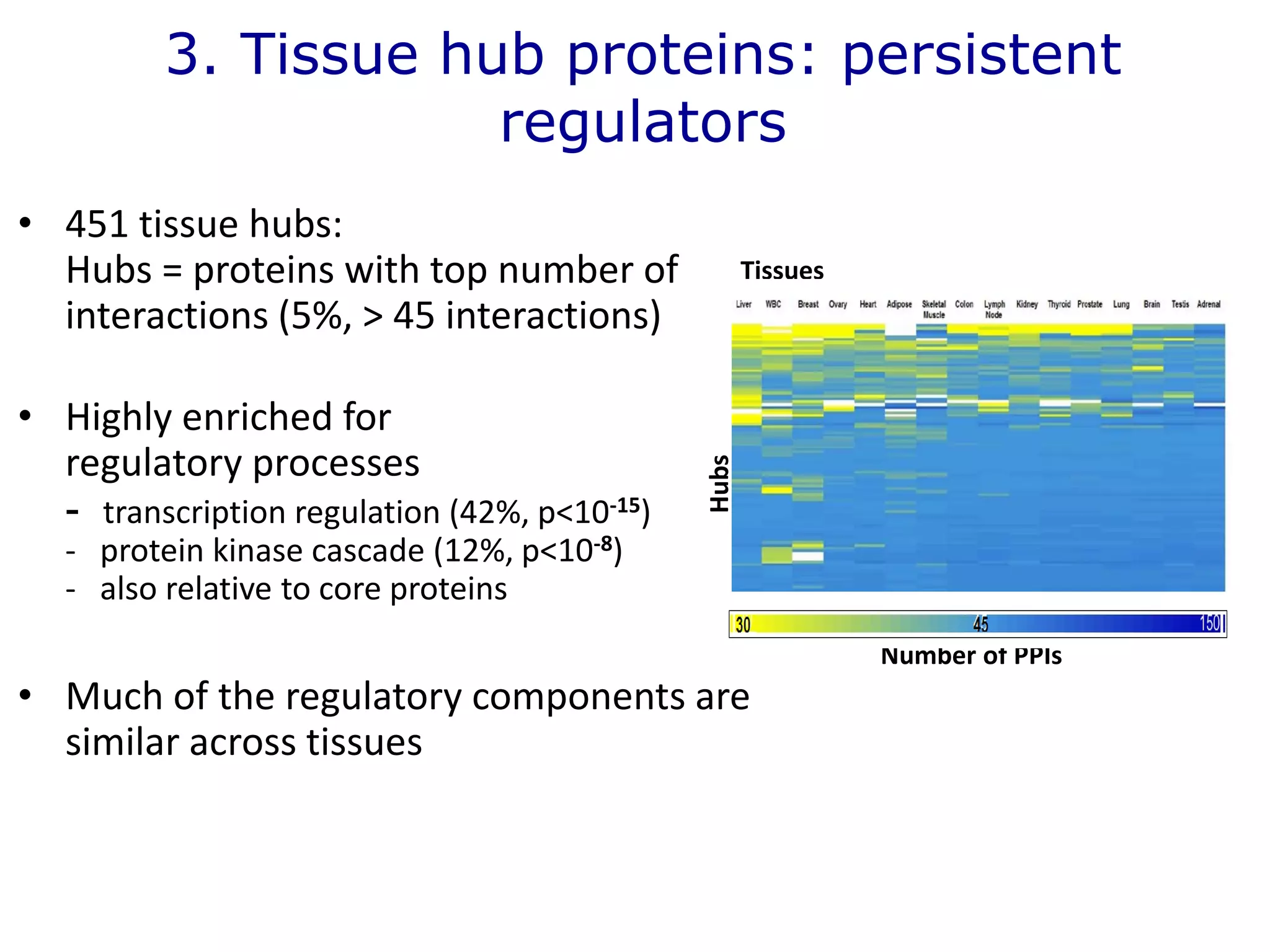
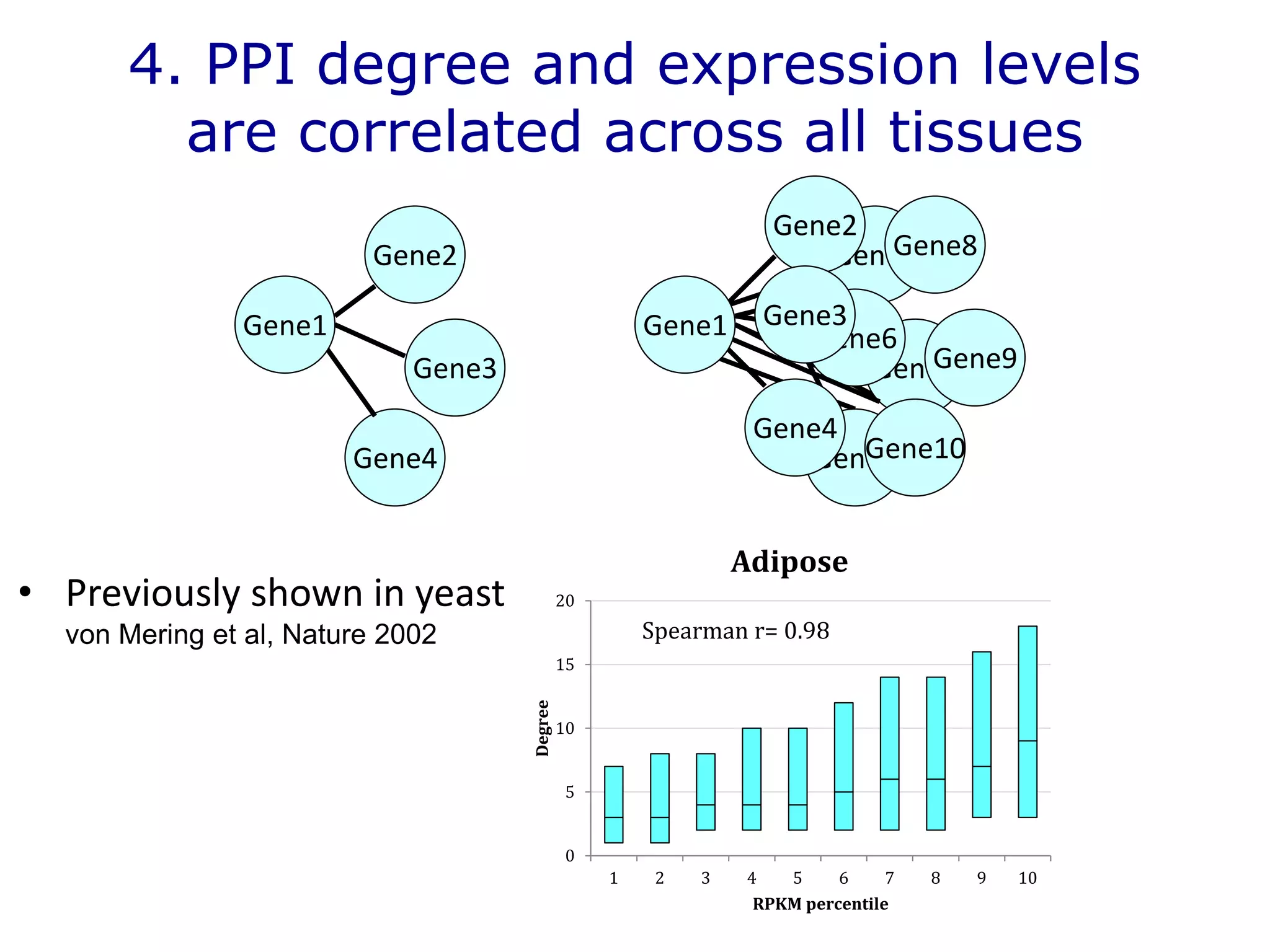
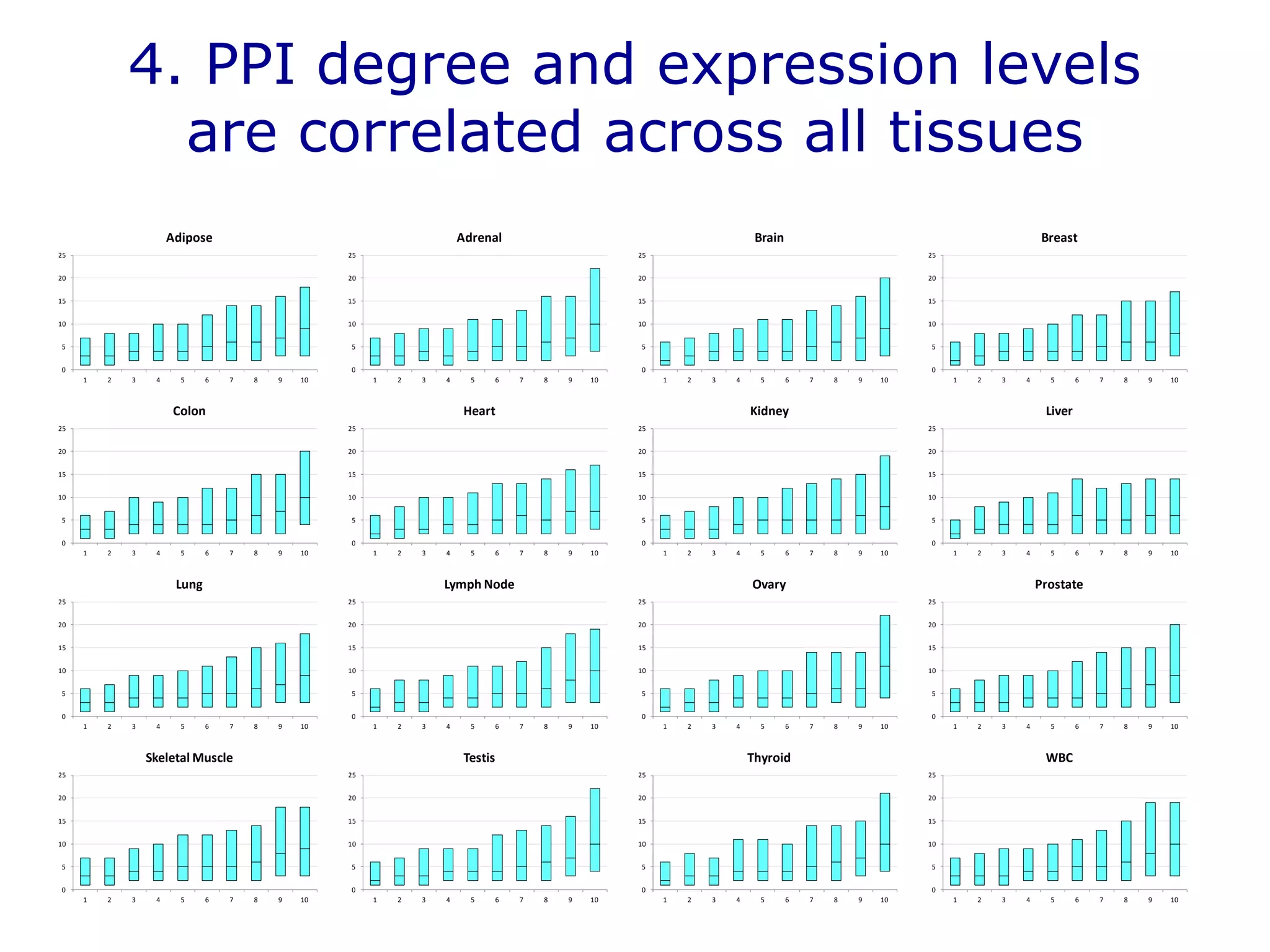
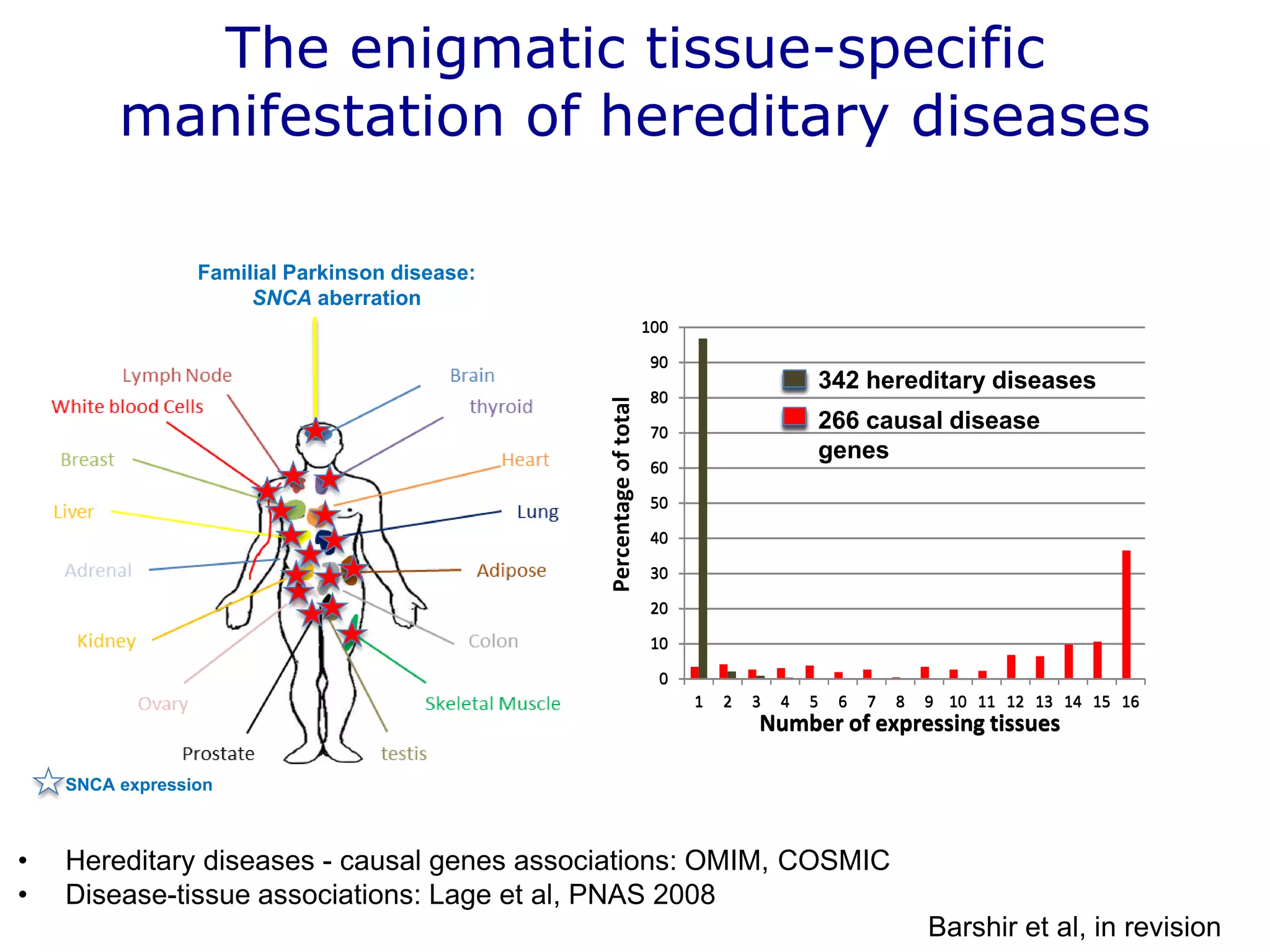
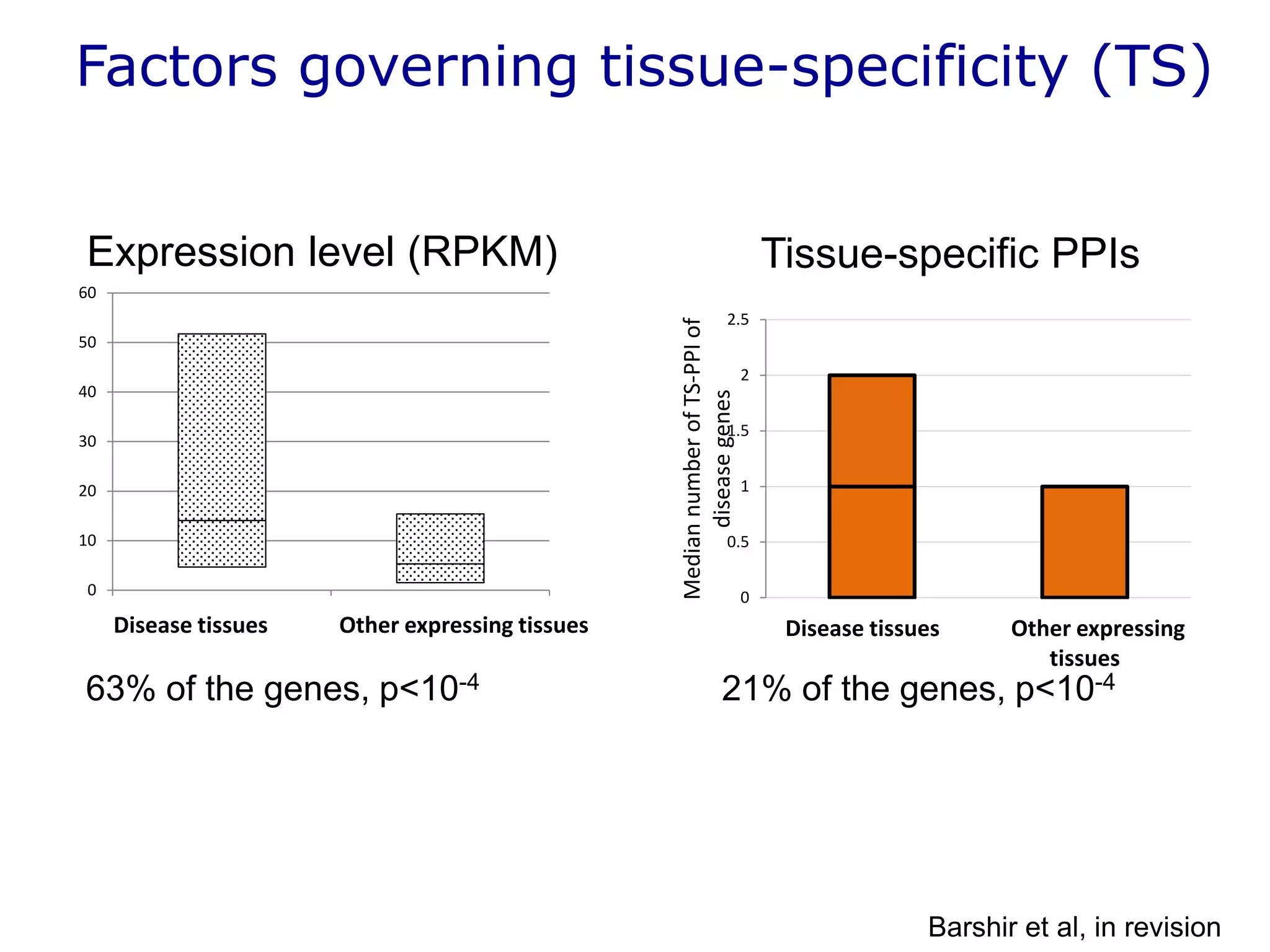
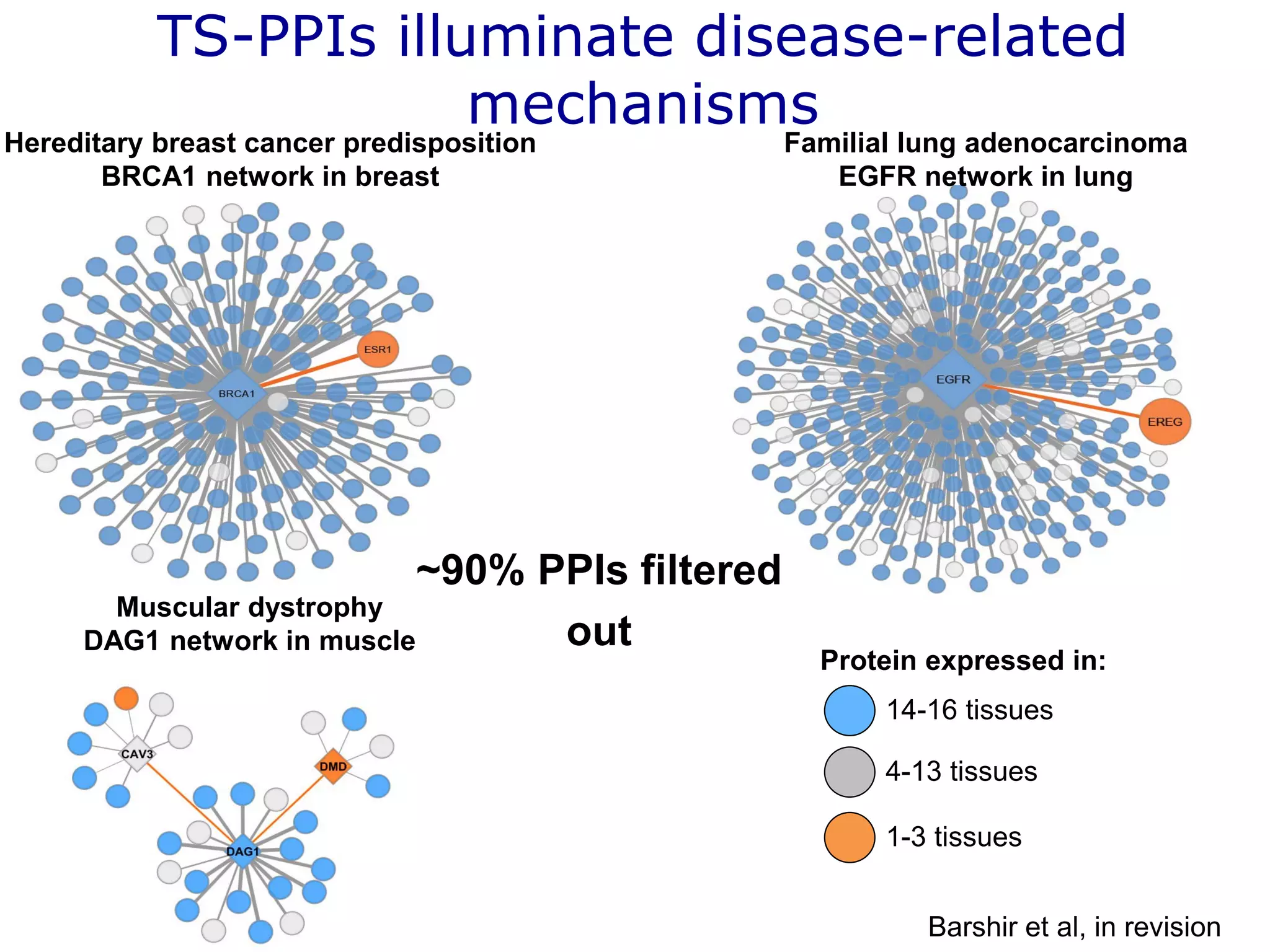
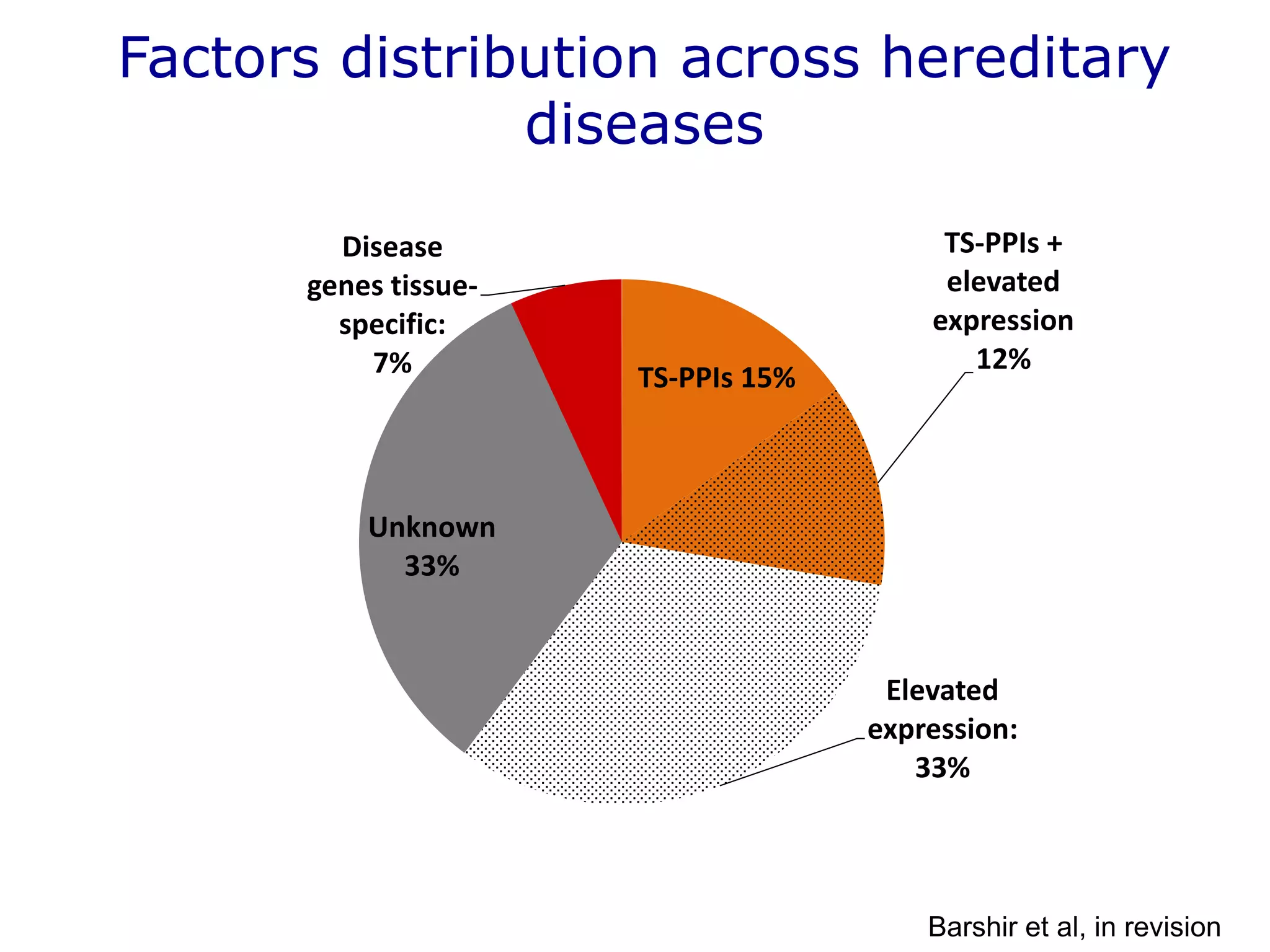
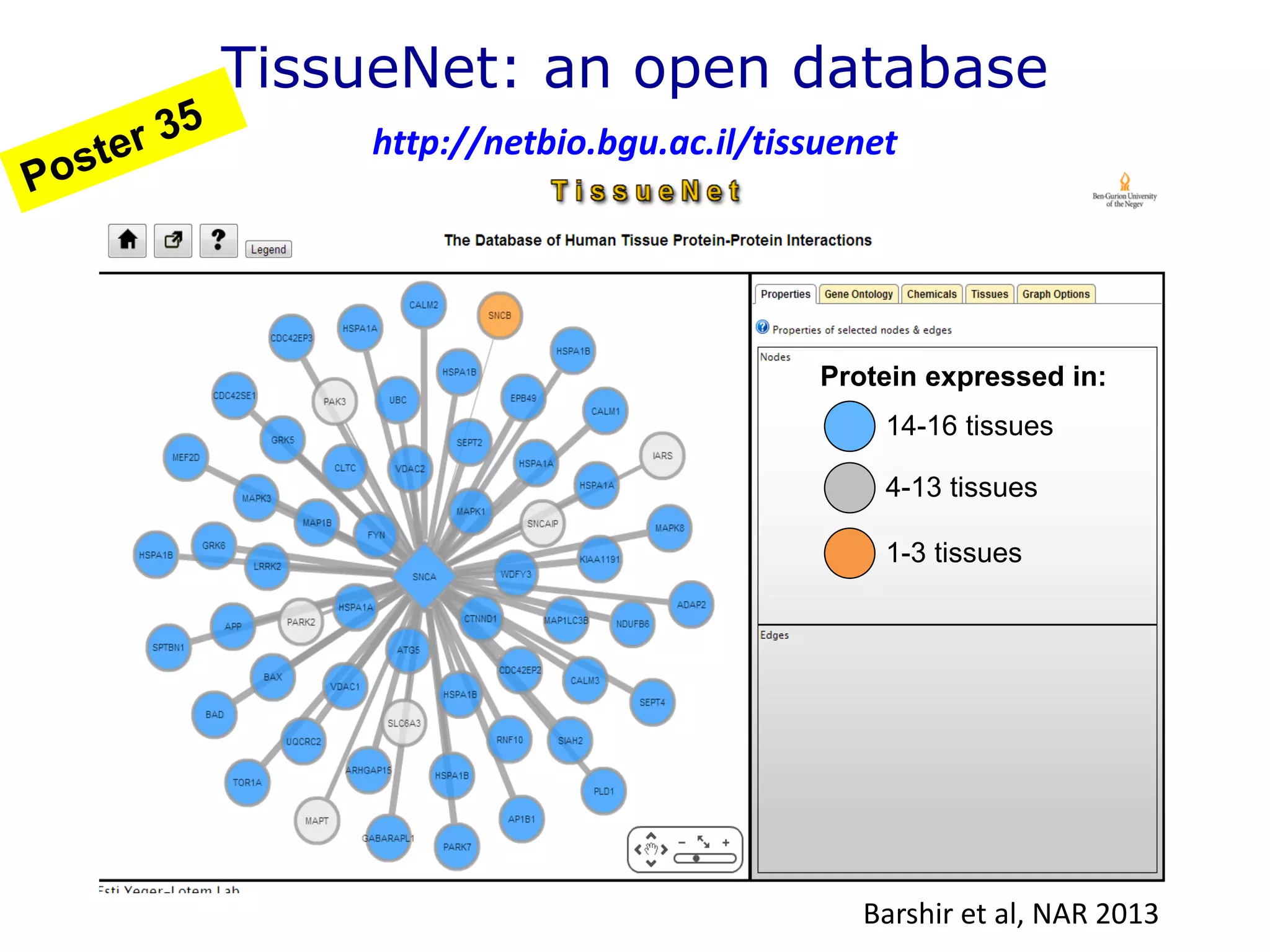
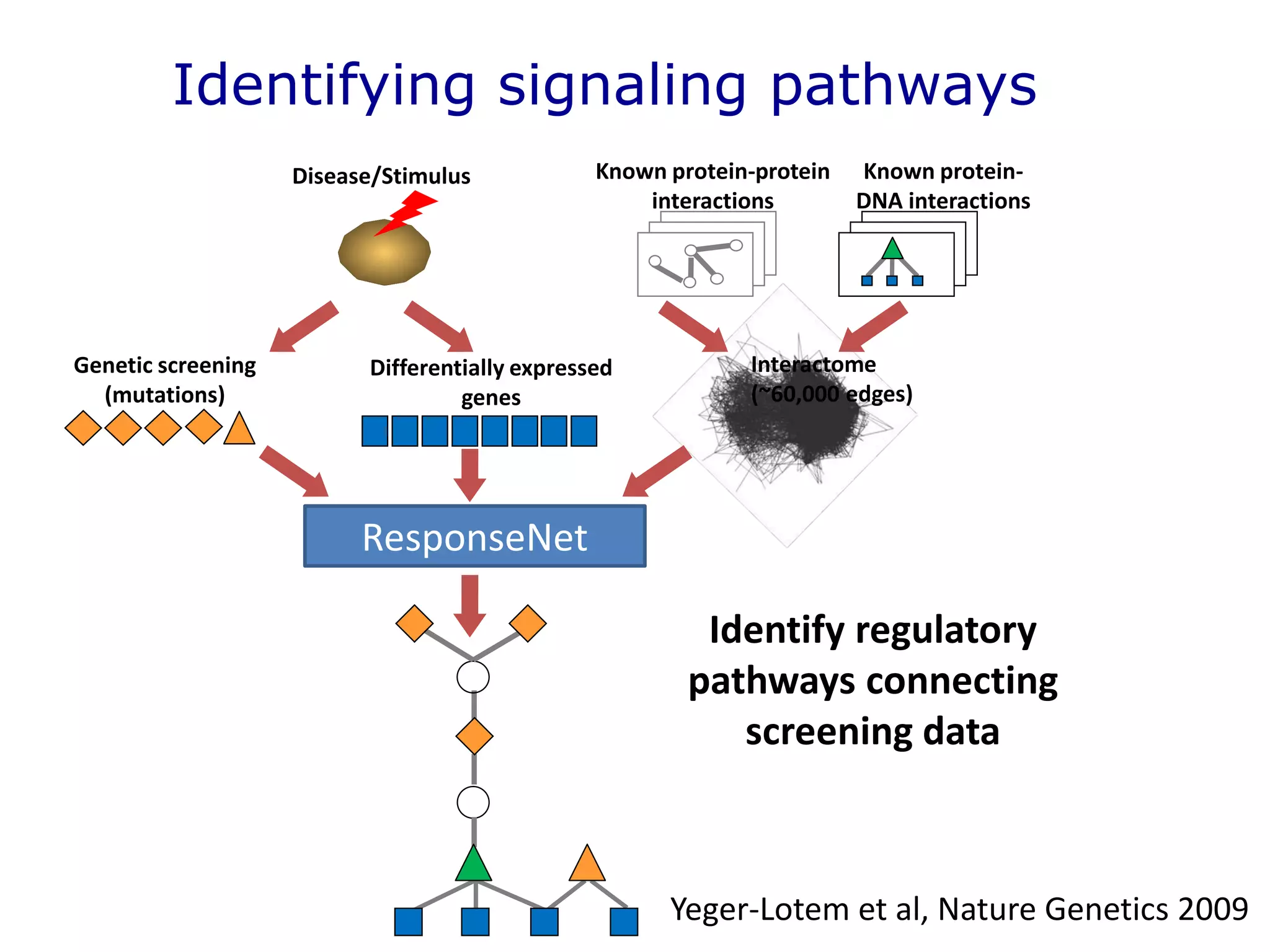
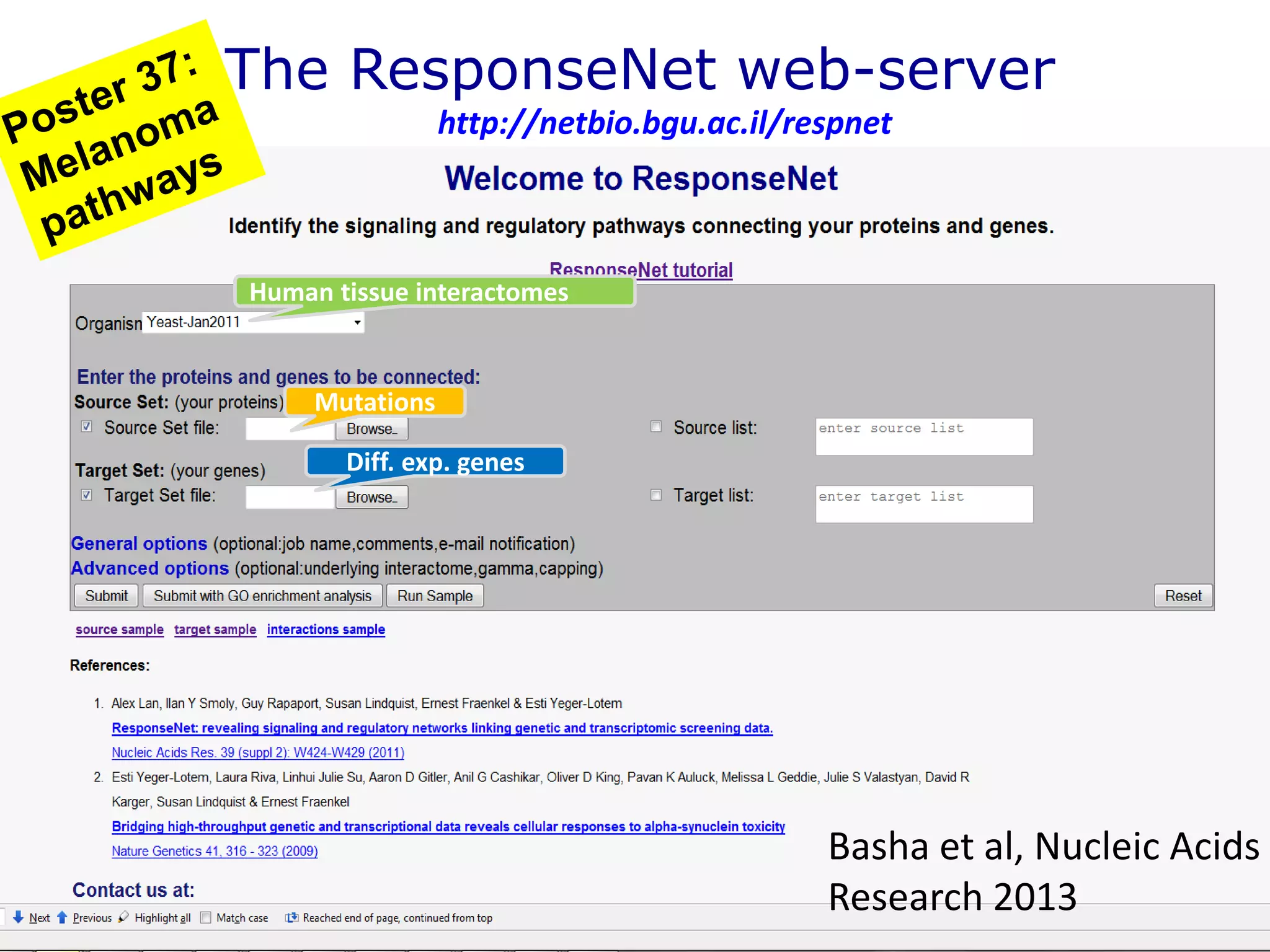
The document details a comprehensive approach to understanding the tissue-specificity of hereditary diseases through the analysis of tissue interactomes, using extensive data from various sources including RNA sequencing. It highlights the development of an open web tool for familial Parkinson's disease and discusses the integration of gene expression and protein-protein interaction data across multiple tissues. The findings suggest that a core network of proteins is prevalent across tissues, and that tissue-specific interactions play a significant role in disease manifestation.