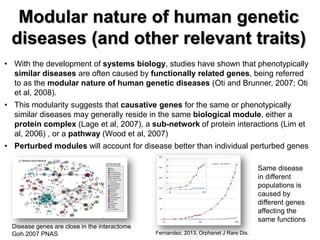
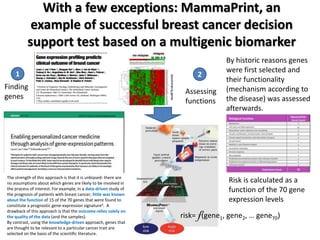
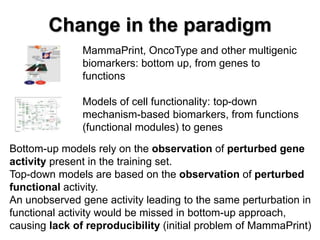
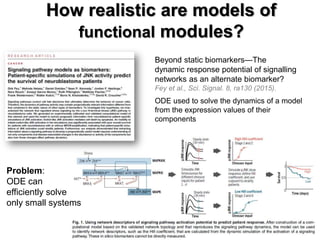
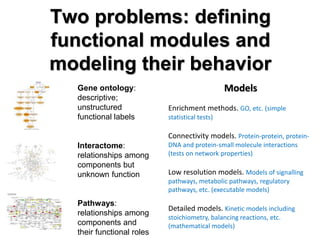
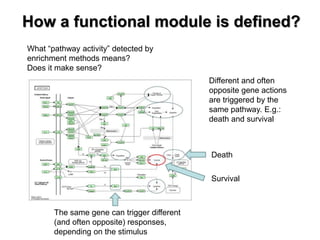
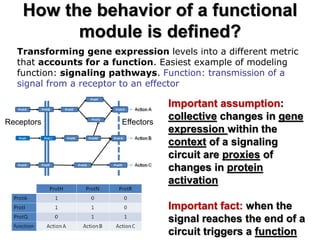
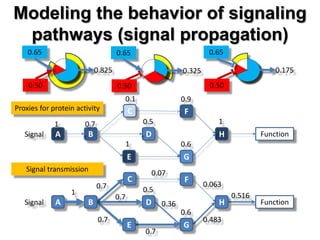
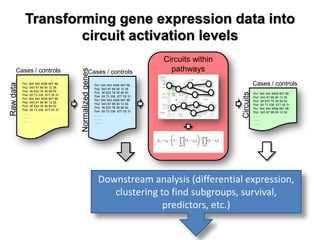
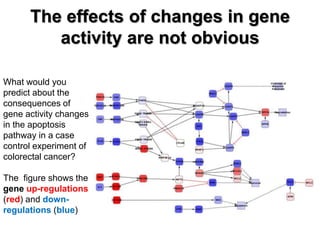
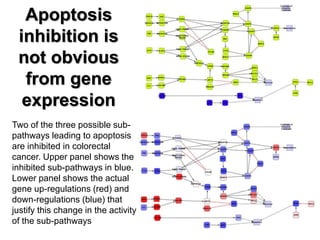
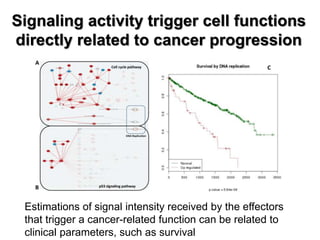
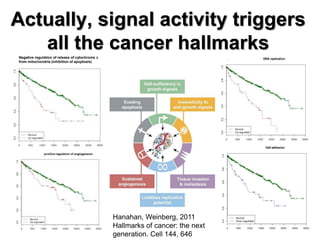
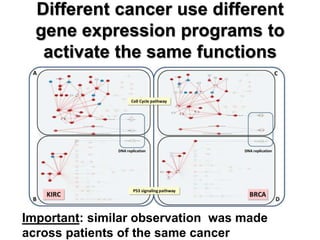
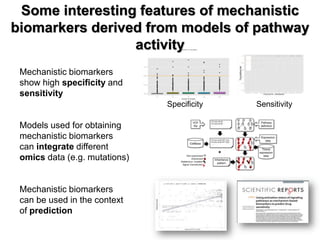
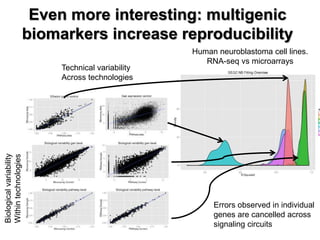
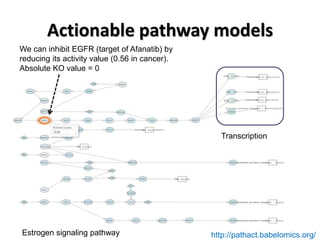
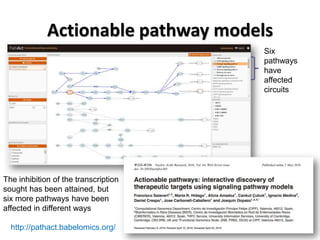
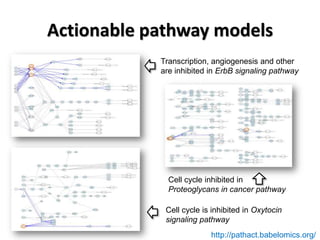
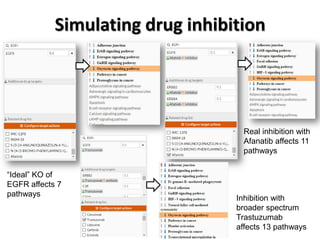
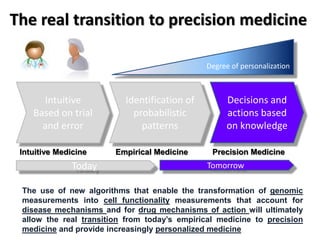
This document discusses the modular nature of human genetic diseases and how disease genes often reside in the same biological module or pathway. It then discusses how most personalized treatments are based on single-gene biomarkers, with some exceptions like MammaPrint which uses a multigenic biomarker. The rest of the document discusses challenges in defining functional modules and modeling their behavior, and how this could lead to more realistic "actionable pathway models" and a transition to true precision medicine.