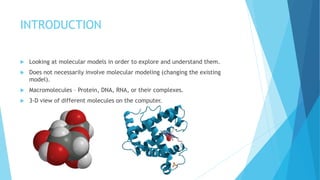
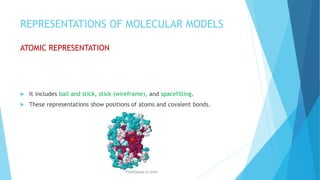
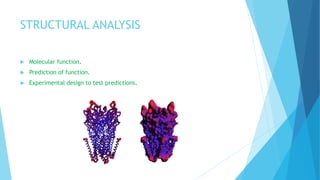
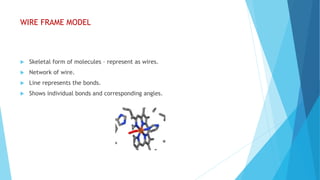
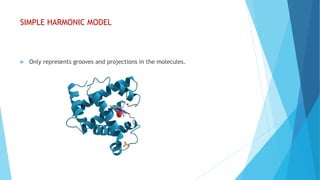
The document discusses molecular visualization techniques used to explore and understand macromolecules like proteins, DNA, and RNA through computer-generated models. It outlines various representations, such as ball and stick, wireframe, and ribbon models, along with tools like Rasmol for 3D visualization. The document also highlights methods for structural analysis and color schemes for macromolecules to help interpret their structures.