1. DNA is made up of nucleotides containing a sugar (deoxyribose), phosphate group, and one of four nitrogenous bases (adenine, thymine, cytosine, or guanine).
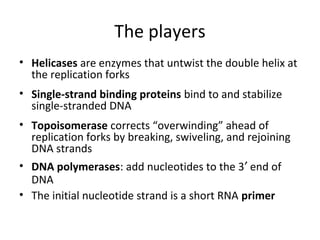
2. DNA replication ensures that new cells have a complete set of DNA by separating the DNA double helix and using each original strand as a template to produce two new complementary strands.
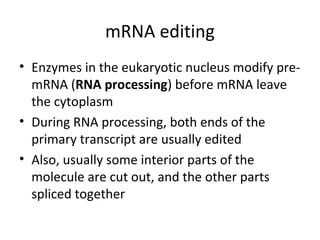
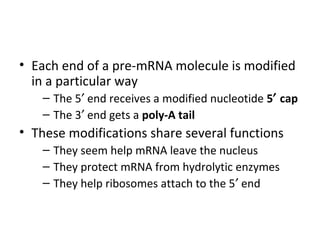
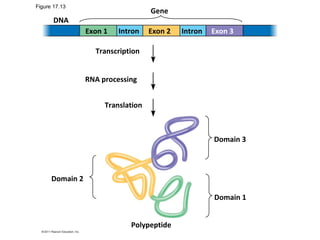
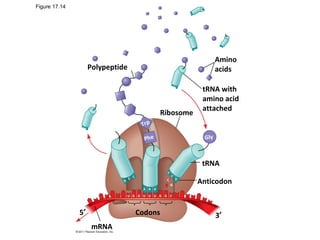
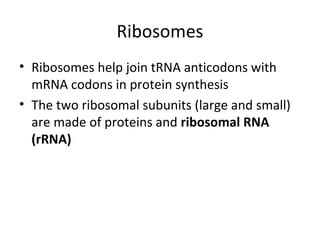
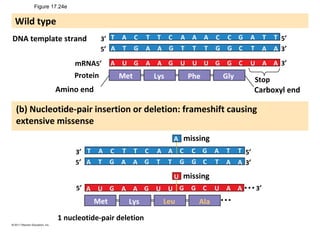
3. Transcription and translation are the processes by which the information in DNA is used to synthesize proteins. Transcription involves RNA polymerase making an mRNA copy of a gene, and translation involves ribosomes using the mRNA to produce a polypeptide chain.