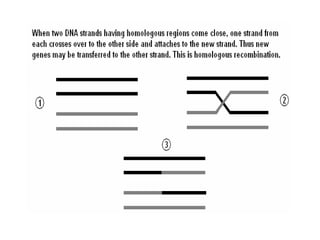
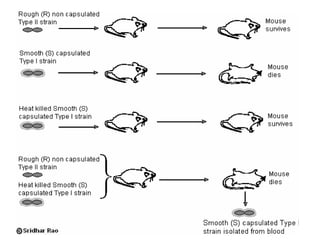
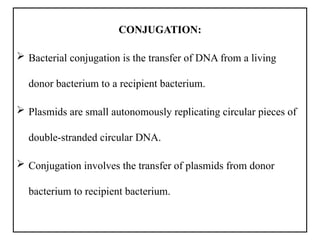
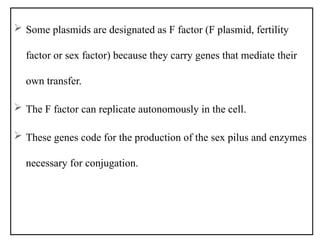
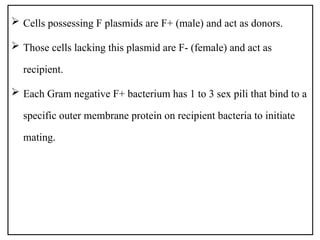
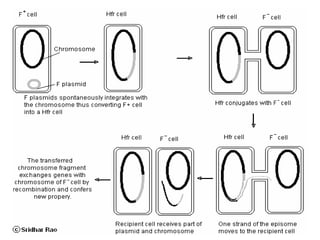
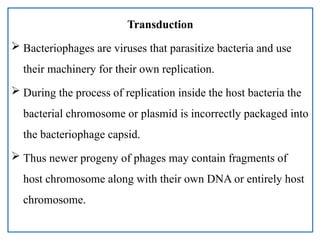
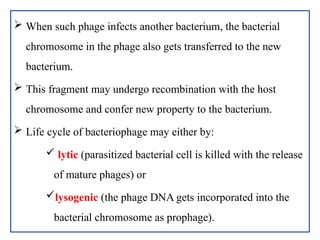
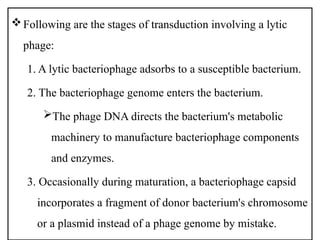
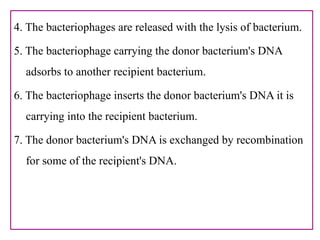
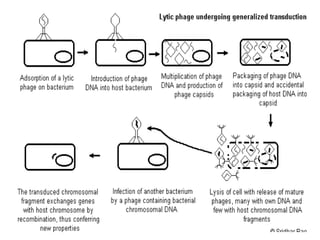
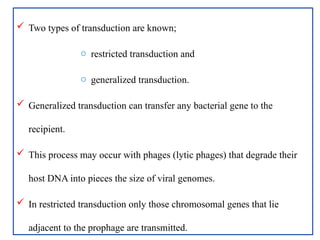
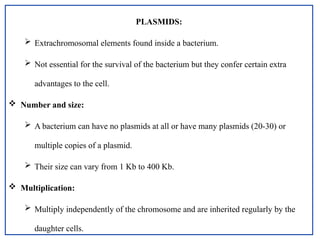
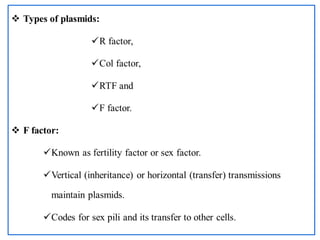
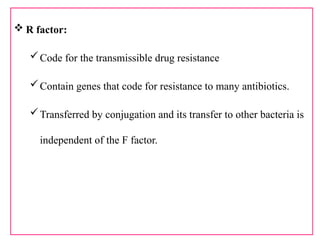
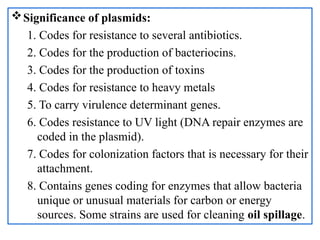
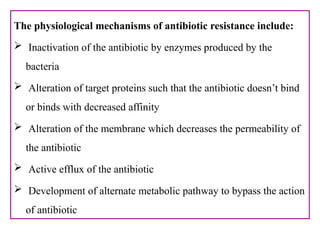
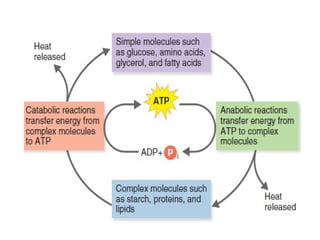
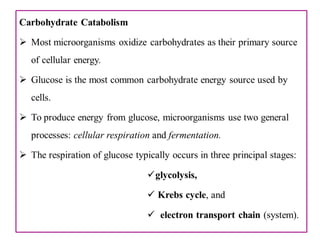
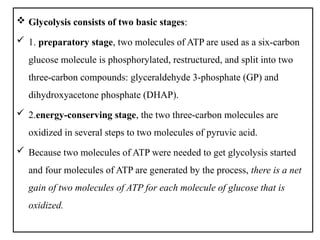
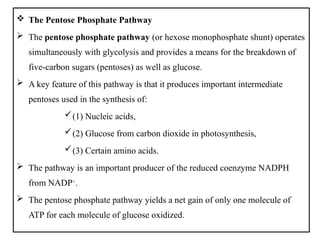
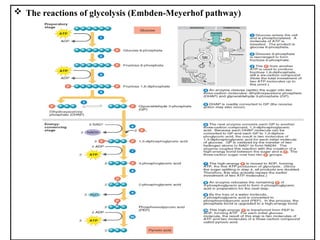
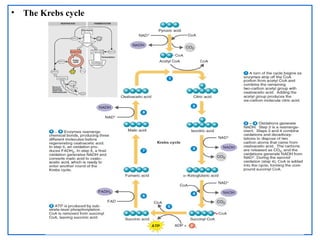
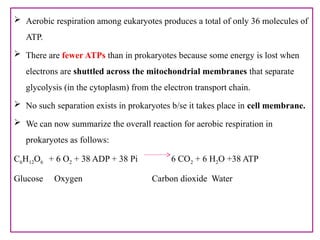
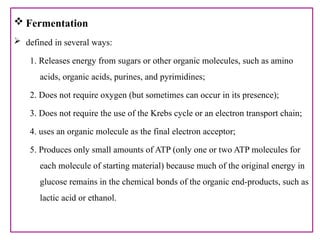
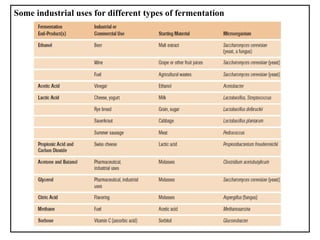
The document provides a comprehensive overview of bacterial genetics, including the structure and function of DNA, the mechanisms of gene transfer (transformation, conjugation, and transduction), and bacterial metabolism processes like glycolysis and respiration. It details the roles of plasmids and their significance in antibiotic resistance, as well as different metabolic pathways utilized by bacteria. Key concepts and processes are documented alongside their implications for gene expression and transfer, contributing to an understanding of bacterial genetics and metabolism.