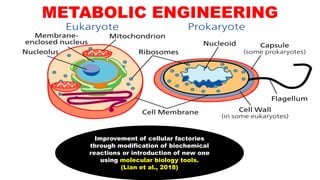
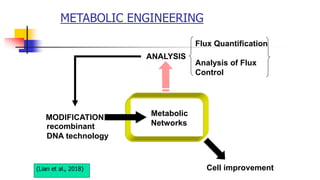
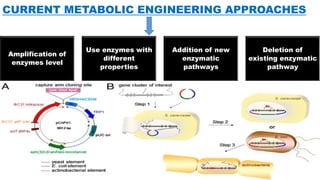
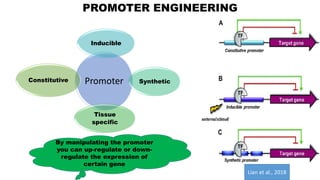
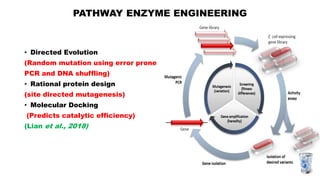
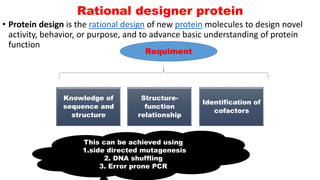
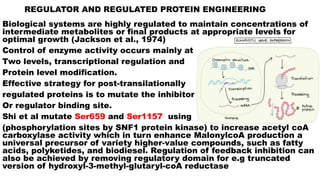
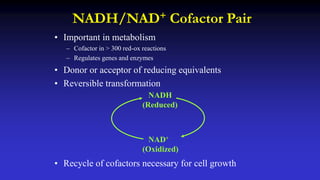
The document presents an overview of metabolic engineering, highlighting the application of biological and engineering principles to optimize biochemical processes. It discusses metabolic flux analysis, methods for modifying metabolic pathways, and various engineering strategies at the biological parts, pathway, and organelle levels. Key focus areas include promoter engineering, pathway enzyme engineering, and cofactor engineering, which are vital for developing new biotechnologies and improving existing metabolic processes.