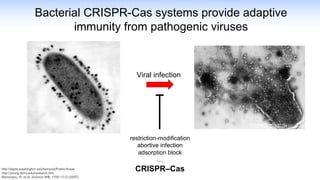
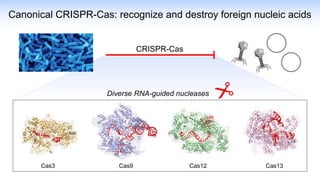
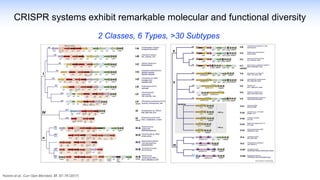
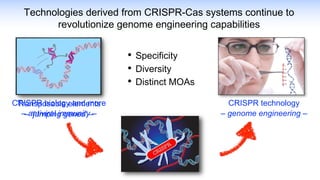
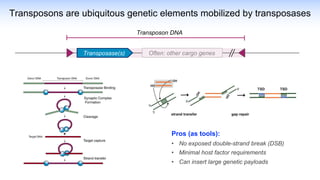
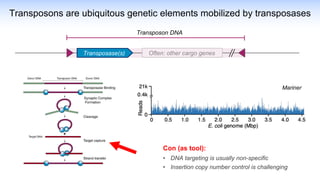
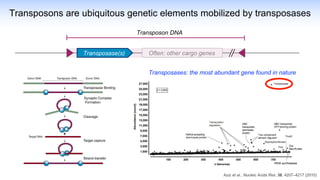
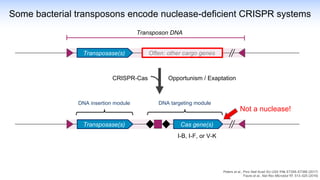
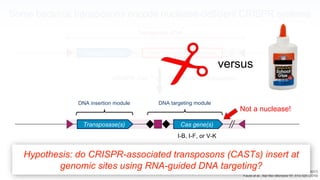
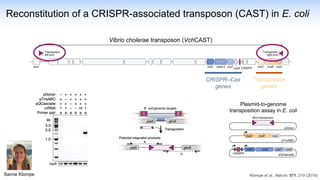
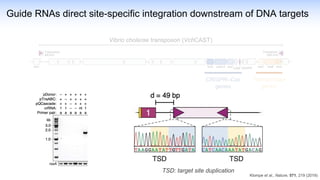
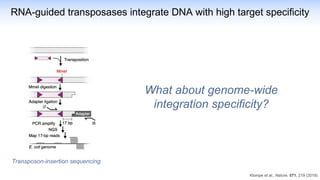
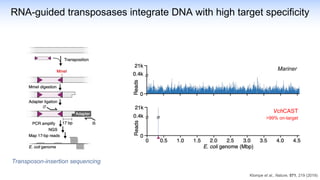
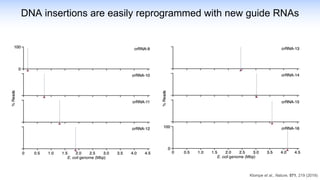
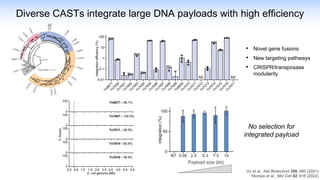
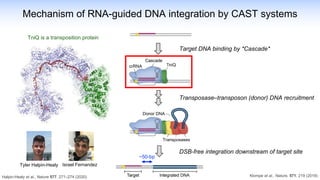
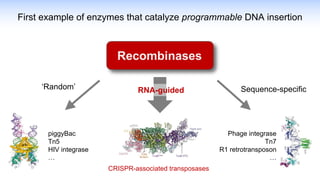
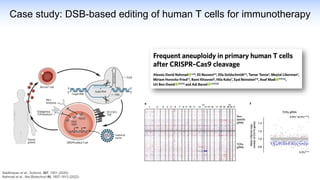
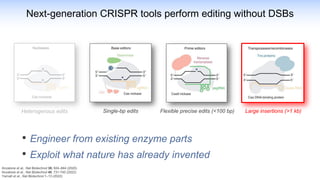
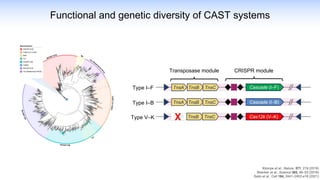
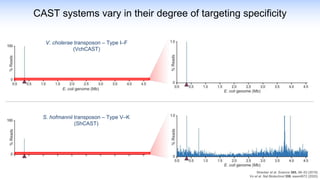
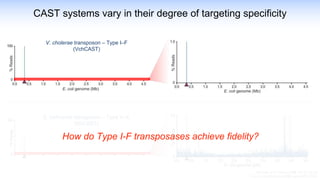
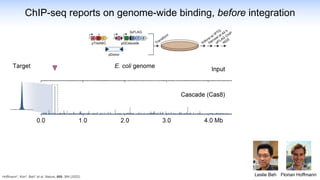
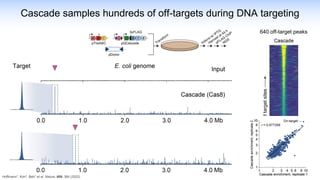
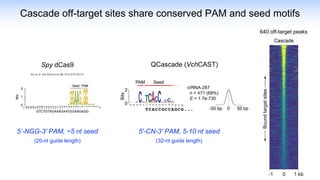
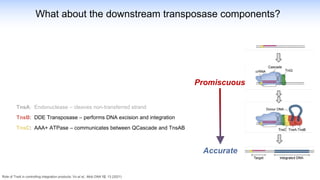
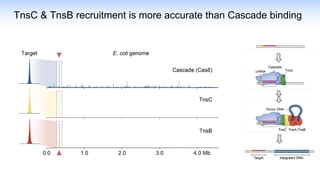
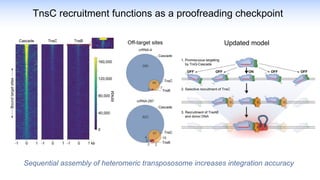
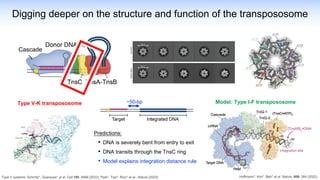
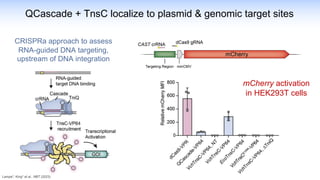
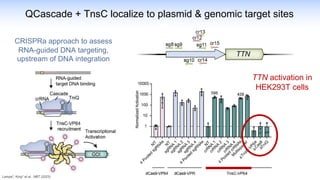
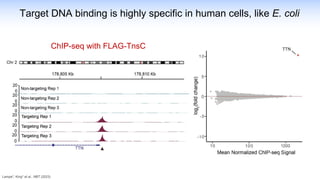
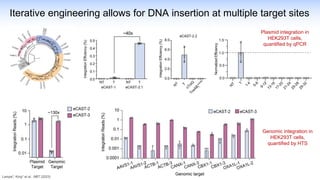
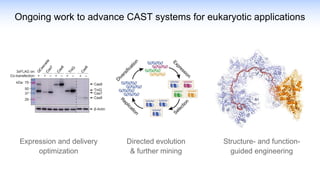
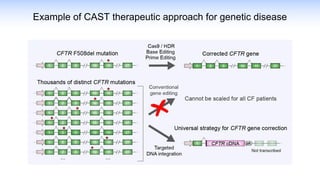
The webinar discusses the use of CRISPR-associated transposases (CASTs) for advanced genome engineering, exploring their mechanisms, capabilities, and advantages over traditional CRISPR methods that involve double-strand breaks. The presentation highlights the diversity and specificity of CAST systems, including their potential for precise DNA integration without causing extensive genomic damage. Ongoing research focuses on improving CAST systems for eukaryotic applications and therapeutic strategies for genetic diseases.