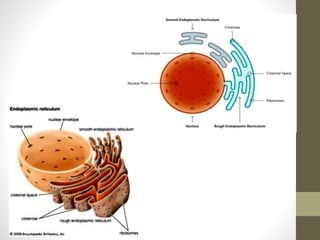
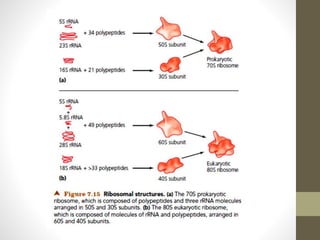
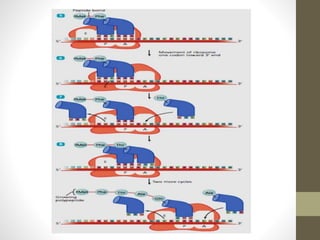
The document summarizes key aspects of endoplasmic reticulum, ribosomes, and protein synthesis. It describes the endoplasmic reticulum as a network of membrane-bound channels found in eukaryotic cells, except red blood cells, and absent in prokaryotes. Ribosomes are sites of protein synthesis and consist of a large and small subunit that bind messenger RNA. Protein synthesis involves transcription of DNA to mRNA and translation of mRNA codons into amino acids by ribosomes, consisting of initiation, elongation, and termination stages.