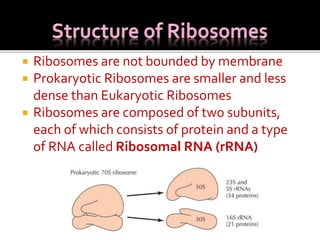
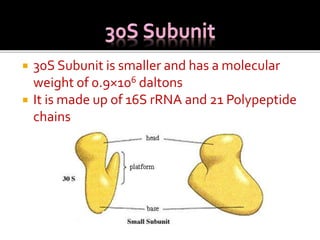
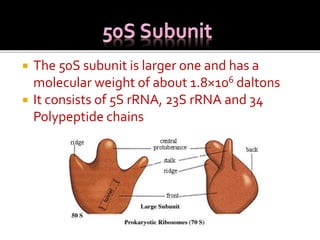
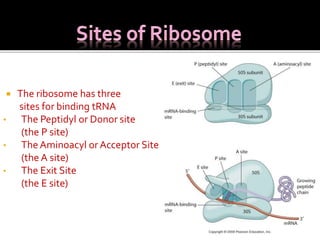
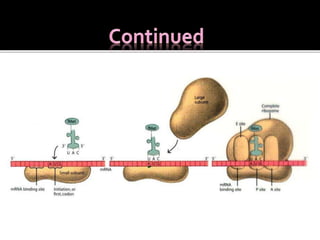
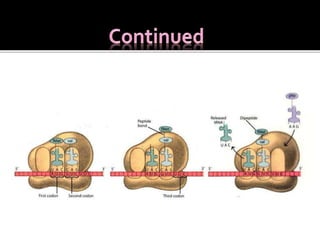
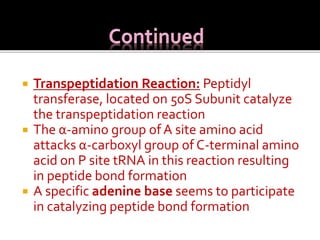
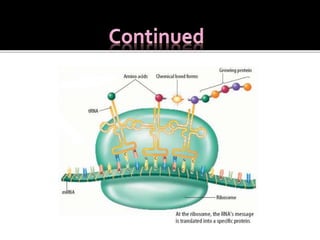
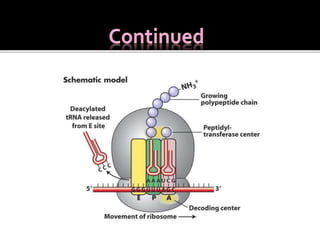
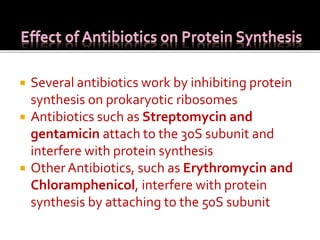
Ribosomes are organelles found in all cells that serve as the site of protein synthesis. They are composed of two subunits made of ribosomal RNA and proteins. In prokaryotes, the 70S ribosome contains a 50S and 30S subunit. Protein synthesis occurs through the three steps of initiation, elongation, and termination on the ribosomal subunits using messenger RNA as a template and transfer RNA to deliver amino acids. Antibiotics can inhibit bacterial protein synthesis by binding to the ribosomal subunits.