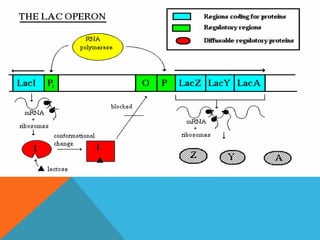
The document discusses the lac operon system, a cluster of genes in E. coli responsible for lactose metabolism. It explains the operon's regulatory mechanisms in response to the presence or absence of lactose and glucose, detailing how the repressor protein and catabolite activator protein (CAP) control gene expression. Additionally, it covers the practical applications of the lac operon in molecular biology, such as its use in reporter gene assays.





































![REFERENCES:
NCBI official website
J.L. Ramos, ... Z. Udaondo, in Brenner's Encyclopedia of Genetics (Second Edition), 2013
Oehler, S.; Eismann, E. R.; Krämer, H.; Müller-Hill, B. (1990). "The three operators of
thelac operon cooperate in repression". The EMBO Journal. .
Griffiths, Anthony JF; Gelbart, William M.; Miller, Jeffrey H.; Lewontin, Richard C.
(1999). "Regulation of the Lactose System". modern Genetic Analysis. New York: W. H.
Freeman.
Koonin E (2009) Evolution of genome architecture. Int J Biochem Cell Biol 41:298–
306 [PMC free article]
Hurst LD, Pal C, Lercher MJ (2004) The evolutionary dynamics of eukaryotic gene order.
Nat Rev Genet 5:299–310
Jacob F, Perrin D, Sanchez C, Monod J (1960) L’operon: Groupe de genes a l’expression
coordonne par un operateur. C R Acad Sci 245: 1727–729
Jacob F, Monod J (1961) On the regulation of gene activity. In: Cold Spring Harbor
Symposium Quantitative Biology 26, pp 193–211
Jacob F, Monod J (1961) Genetic regulatory mechanisms in the synthesis of proteins. J
Mol Biol
Warren PB, ten Wolde PR (2004) Statistical analysis of the spatial distribution of operons
in the transcriptional regulation network of Escherichia coli. J Mol Biol 342:1379–1390](https://image.slidesharecdn.com/lacoperonsystem-210818040658/85/Lac-operon-system-39-320.jpg)
