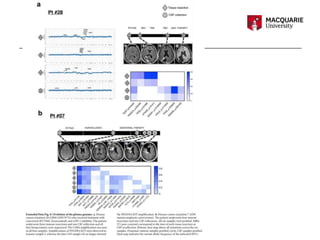
Tumour DNA was detected in the cerebrospinal fluid (CSF) of patients with glioma. Analysis found the CSF tumour DNA provided a representation of the tumour genome that was genetically consistent with tumour tissue. While the study was limited by a small sample size and selection bias, it demonstrated CSF analysis may be a reliable way to determine the glioma genome and could provide prognostic value. Further research with larger studies is needed to validate using CSF genomic analysis in glioma patient management.