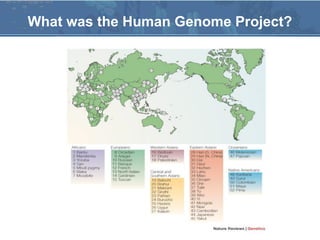
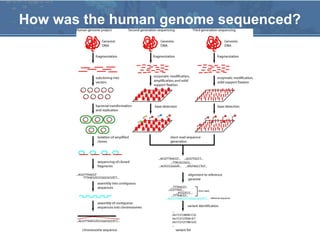
The Human Genome Project (HGP), conducted from 1990 to 2003, aimed to map and sequence the entire human genome, providing the data freely to the public to enhance understanding of genes and disease. The project involved collaboration between 20 institutes worldwide and utilized advanced sequencing technologies, completing the initial draft by June 2000 and the final sequence in April 2003. Subsequent projects, such as the HapMap and 1000 Genomes Project, continued to explore genetic variations and their implications for health.